













| General Information: |
|
| Name(s) found: |
mug87 /
SPCC1620.11
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 851 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA cytosol [IDA nuclear pore [ISS |
| Biological Process: |
nucleocytoplasmic transport
[ISS poly(A)+ mRNA export from nucleus [IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVASDDSPK EARGIPFLDQ KSRKLANELL EPCLPFIQFN LGEIEQRAKH YLNTVPTSKD 60
61 GNTKAHYLLA GSGINAEQTW KKIESLSLQV RPPTTLELSF TDVDMFLKYH REKNVLNSLE 120
121 ALVQNTQIAF DQYLEEEWRS KAAKSRPSFD NILLENKKRV SFYPFSVQRS QKFASTLKMC 180
181 LEEEALHGFQ SKLVSSFCEV AREFAHDTKS LLLYESWKLL SSVILDKDSV TVFGNKGIIS 240
241 KAFDIETEDG SVNSRFYQRI SDCSRKFLEA QFFEVLNKEI AKTPQAALVG GVPSIRNKIR 300
301 AYLNIRLLRN GVWINPDLEI IQDVPIWAFI FYLLRCGFLK EAVDFTEENR DLFEKVAEKF 360
361 PFYINAYAKA PNGILPRQLR SQLFSEFNQT IRLQESSDPY KYAVYKIIGR CDLSKTSCPS 420
421 ICSVTEDYIW FQLILSREFT EKSVSAHEFF SLEDVQHILL SYGSDYFTNN GSNPVMYFFL 480
481 LMLCGLYERA INFLYPYFPT DAVHFAITCA YYGLLRTAPS SSVVSNEPGK IQSMLVETKS 540
541 GKPSLEFDRL LIDYTQTCQE LSPVMSACYL IPMCKIDKYI SMCHKSLCSL VLSTRDYVNL 600
601 LGDIRGDGER TPSFLENHRS LIGLSSVKEY LSKITLTAAK QADDQGLLSD AILLYHLAED 660
661 YDAAVTVINR RLGSALLRFL DQFVFPDKLI SLTKSMMDVY NRNPSLYAKV DYKNRETTNL 720
721 LLLTVEAFNA YTNKDYEQAL SSLQQLEILP LDPLDSDCET FVVRKLAKEF RFLNENLLQN 780
781 VPGIVLIAMN SLKELYAKQK SSSFGNDAIS VDKLRLYRQK ARRIVMYSFL IEYRMPSQIL 840
841 EQLNRCEIEM T |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
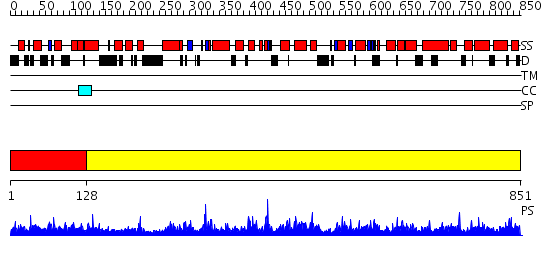
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..127] | 2.036995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [128..851] | 1000.0 | No description for 2qx5A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)