













| General Information: |
|
| Name(s) found: |
tas3 /
SPBC83.03c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 549 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mating-type region heterochromatin
[IDA nucleus [IDA nuclear chromatin [IC nuclear telomeric heterochromatin [IDA RITS complex [IDA spindle pole body [IDA centromeric heterochromatin [IDA |
| Biological Process: |
regulation of histone methylation
[IMP meiotic chromosome segregation [IMP regulation of chromatin silencing at centromere [IMP chromatin silencing at silent mating-type cassette [IGI cellular protein localization [IGI chromatin silencing by small RNA [IMP |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKGIKKYLP SLPFLACISD FPENHGTSRR SATVSLERVH ELFTEHWLSN LKNRREKRQE 60
61 LAEEAVYCRS EMLSQRKLLA AVDFPQQLKN SPAKAKATHT SSGVTKEVRA SKKYTSSNVE 120
121 FPLVTDGKEK PVKSKQLRKN SVTEFEKPIE TKKSKHRKSR NKFLDKSSGS MEIESWDNST 180
181 SDSIIESSSR LHESISLREN DIRSSDSKSV GWDDNSTGFR ESSKSLDHTD TSMFMELDSN 240
241 SDPQFRPKYQ AKSSWFAPDD PEASWGNLDD GWGETNGSWS STDDTKHYKN EWAESINLDN 300
301 FNRPSQQEDY DKPKNTQVSR SSNHHRRYDS YHPDSRSDSY RSKREHYDNR DTGPRSKHLE 360
361 KSSYVYNQNF EDRTHLSDHG AHFHLGNAND FNMQGSSRKR KASDRQRESR ENELPTKKLN 420
421 ASDSHNPLAS LTTDKNDLYI NWLKSLSFFQ TNSSCAEALV KVIPHYHNKL IDFSQVLQLV 480
481 FSASEKFPIQ ENQPLPEQLM FLSNLEKQTP FAKAVGSSIY KLVTGKNLSL DFASQILKEA 540
541 SILEHKNEK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
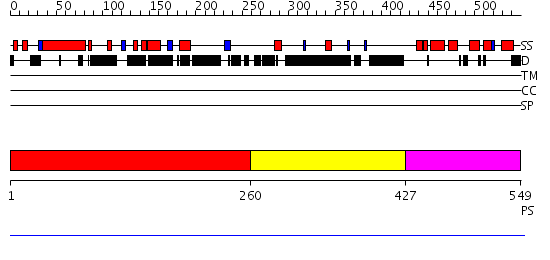
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..259] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [260..426] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [427..549] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)