













| General Information: |
|
| Name(s) found: |
SPAC644.16
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 422 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mRNA cleavage factor complex [ISS |
| Biological Process: |
mRNA polyadenylation
[ISS mRNA cleavage [ISS |
| Molecular Function: |
RNA binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNPSCVVYVG NIPYEMAEEQ VIDIFKQSGP VKSFQLVIDP ESGQPKGYGF CEYHDPATAA 60
61 SAVRNLNNYD AGTRRLRVDF PTADQIRRLD KLLGPSRYGY YPQSYANQSY TYGNNFGSYP 120
121 PTQPSTQPLP QSYGYPSYPP AGYRGGSARP SGVLANDEVY RVLAQLAPNE IDYMLSAIKA 180
181 LCLEAPEQAA QLFETNPQLS YAVFQAMLMK RYTSESVVAD LLIPAGVNLP GAQEPNRGYF 240
241 SPMHTYSSAV PGPISVPSAP YGRASSTIAE VSPMYGSHAA PYASTPSAAV GSSRGSTPAS 300
301 ATVPISPARG FPTTSAYNPA PPAYGMANPA YGSTGIRSSS IPSSGSIRSP SLTTTSAQAT 360
361 TNATNNITTT TAAQDENATK AALIAQLMAL TDDQINVLPP DQKERILQIR QALPSSYKTE 420
421 SK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
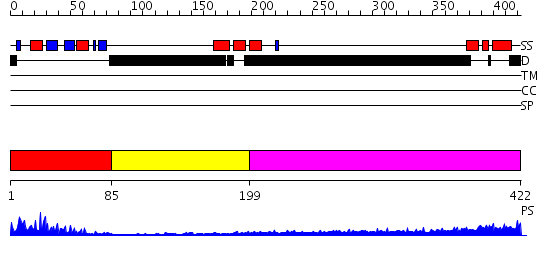
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 19.0 | NMR Structure of the N-terminal RRM domain of Cleavage stimulation factor 64 KDa subunit | |
| 2 | View Details | [85..198] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [199..422] | 5.522879 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)