













| General Information: |
|
| Name(s) found: |
SPBC23G7.10c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 395 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
generation of precursor metabolites and energy
[RCA cellular response to stress [IEP |
| Molecular Function: |
FMN binding
[IEA]
NADPH dehydrogenase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTIVNEGAEN VGYFTPAQKI PAGAAIGVPQ TKLFTPLKIR GVEFHNRMFV SPMCTYSADQ 60
61 EGHLTDFHLV HLGAMGMRGP GLVMVEATAV SPEGRISPND SGLWMESQMK PLRRIVEFAH 120
121 SQNQKIGIQL AHAGRKASTT APYRGYTVAT EAQGGWENDV YGPNEDRWDE NHAQPHKLTE 180
181 KQYDELVDKF VVAAKRAVEI GFDVIEIHGA HGYLISSTVS PATNDRNDKY GGTFEKRILF 240
241 PMEVVHSVRK AIPDSMPLFY RVTATDWLPK GQGWEIEDTV ALAARLRDGG VDLIDVSSGG 300
301 NHKDQRIEVK DCYQVPFAEK IKDQVNGILL GAVGMIRDGL TANEILESGK ADVTFVAREF 360
361 LRNPSLVLDS ANQLGENVAW PVQYDYAVKG HRKLR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
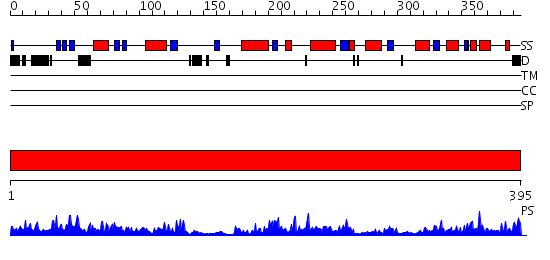
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..395] | 90.09691 | Crystal structure of oxidized YqjM from Bacillus subtilis |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)