













| General Information: |
|
| Name(s) found: |
zta1 /
SPCC1442.16c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 329 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
mRNA metabolic process
[ISS |
| Molecular Function: |
AU-rich element binding
[ISS zinc ion binding [IEA] NADPH:quinone reductase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNLLVQVSK TGPSSVLQVI TKEIPKPAPN GLVIKNAYAG LNYIDTYLRT GLYTAPLPYI 60
61 PGKEAAGVVA AVGDKVEADF KVGDRVVYLT PFGAYAQYTN VPTTLVSKVS EKIPLKIASA 120
121 ALLQGLTAYT LIEEAYPVKT GDTVVVHAAA GGVGLLLCQM LRARNVHVIA TASTAAKRRI 180
181 AIKNGAEIAC SYEDLTKVVA DYTNGKGVDA AYDSVGIDTL SSSLDALRNG GTMVSFGNAS 240
241 GAIDAIPLKF LSARCLKFVR PSLFGYITGH AVFEGYVSRL WKEILDNNLN IAIHHIFKLS 300
301 EAKEAHDAIE SRATTGKLLL LCNEDLADA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
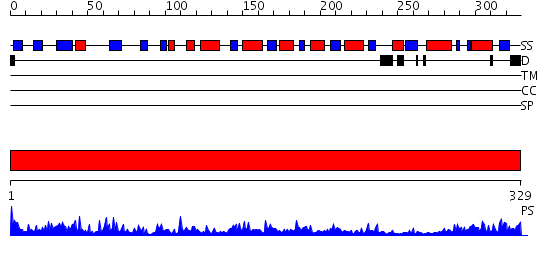
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..329] | 63.39794 | Quinone oxidoreductase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)