













| General Information: |
|
| Name(s) found: |
SPBC354.10
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 963 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
transcription-coupled nucleotide-excision repair
[ISS telomere maintenance [ISS response to DNA damage stimulus [ISS ubiquitin-dependent protein catabolic process [ISS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSENKKFRPK KVGNTEQVSE KQHTDTALNI SQDNKTSSSS AKRGAKGRRG TKGKSGPANS 60
61 SQTSNIEGSN ALELEFDRDS QLSVLQELFP SWTIDDLSFA LEEADRDLEL TILHITEGHA 120
121 SKWGEVKKKP IPKTKSKPTS HAPVSDNVSS TFRNATRKSK KPSASKDTSR GVRKSKAGAP 180
181 SDPSSVHAPS SLEKPAGTGD LPSSEISTKA PASTTVSSSV DPGTINEDSS MKDHTTSNEQ 240
241 ASVLTSANTA ASTNTNGTAG GTSASAKSTS AADQAVASSK PIKKAWASVA KSKKKVTPAP 300
301 APAPESEPSK PSIAPSQPSK TNVSAAYEKP AELSSSSVPF PHKSQDSATP ANVETTPSTA 360
361 TSAPKKSTAP FAINAVKPAP GLSNISSASL PKPSFAKQAA VGSQSSTTSM TTARLLSDRF 420
421 PVVMPVANTA AIPEKVQVRF GSLTLGGEDK KSTKSSSDNI AQSGPRSSYF PKKTVSPKPE 480
481 AKKEASKVAE STKIPKKQHT SAYESRAPQS KVPENLKESH VNETPYRGLH DVNLPASGNA 540
541 SSVSAIPPQV SAQTTIPAAS TSATSGPVVG KQSAGYGGIY SPATHGISPQ PYAGASLIPP 600
601 STGAAFNNET ASSNAGETTA LPYSSRFMNP VENRSANSPF DNFQHIDHST ELGYASNTSQ 660
661 QYQQTSHPAY TTMSPIEYGQ AAPAAHYYNE RNQYSQYDNV AGSASNTPDV HSVMHNKVAS 720
721 SNATSLPAAG TATPSPVVSQ QQPQPYAFPP MYPIPYVSYG YGTMPYNNNK FGQPQQGYMS 780
781 QSGFNDFPPI FGGHSNVYNR QQPGNVSGMS GTQTSNPINN ATANTSGMTE KAAGHRDSVI 840
841 GNGGVTGANS MSSTLGGLSS MSGMGRAAPG MFMGNVGSFE NLSANAASPY GLPPQQTPLT 900
901 NAATQQTTSF QSNANKVNNA NSQPAGFPFN SGASTNAGSN GLGGYNAYNQ SFINRPGGWY 960
961 GNA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
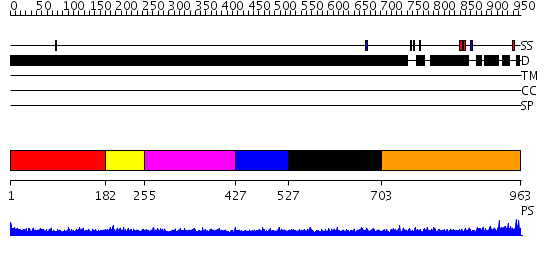
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..181] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [182..254] | 2.17899 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [255..426] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [427..526] | 1.183993 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [527..702] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [703..963] | 3.07 | Crystal structure of the C-terminal domain of BclA, the major antigen of the exosporium of the Bacillus anthracis spore. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)