













| General Information: |
|
| Name(s) found: |
SPAC6G10.07
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 780 amino acids |
Gene Ontology: |
|
| Cellular Component: |
commitment complex
[ISS nucleus [IDA nuclear cap binding complex [ISS perinuclear region of cytoplasm [IEA] spindle [IDA cytosol [IDA |
| Biological Process: |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay
[ISS nuclear mRNA splicing, via spliceosome [ISS |
| Molecular Function: |
mRNA binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSYRGSTRP RKRTREGENY GFRPHRGNSQ ELLAARIKKD ITFLADPRGN SVAADDINYV 60
61 AMSLSREAND PETISTILDC IQTTAFIIPV KIPHLATLII RASLRVPLIL EKAAAYFCLQ 120
121 YFTNLNSFLY YEAKVDLRML ICMSFALQPG TLKPLFSLLA DAISKETKPS VWGDNFLRII 180
181 LINLPYFIAA NNDLGKKDFA NEILDQCEIY VRHRKSSITL SNPLSIHDNL SEEELDLLYK 240
241 QLILSRENDF TFPYISQPWK FFESDFVHIV PVSPSIPEWT FQPTPQQNEL PSFKRFFELF 300
301 NNFEIRTTPD ASDVAASIFR DISVDVINHL EFNRVEAAQV LTDLDVYFTY KTFALRGTPV 360
361 NELPNLDPSE SRWKAEDIIV EAVLGELLGS QNTTYKPVYY HSLLIECCRI APKILAPTFG 420
421 RVIRLMYTMS SDLPLQTLDR FIDWFSHHLS NFNFHWKWNE WIPDVELDDL HPKKVFMRET 480
481 ITRELILSYY TRISDSLPEE LRCLLGEQPS GPNFVYENET HPLYQQSSQI IEALRLHKPL 540
541 EELDIILQSE EIQNSETSAV RLVMSCAYSL GSRSFSHALN VFEKHLNTLK HFSRKSLDSE 600
601 IEVVDELFSF WKLQPFNAVM WLDKMLNYSI ISITSIIEWL IKQDVTIWSR SYTWSLVNTT 660
661 FNKLAARLRR SVSNKEDSSL INEANEEKEI VTNLLLSALR ALISENAENI WVSHWLNLML 720
721 KYVESNFLSV KKDTIEEANE PVQENTSEEQ EDTKMQPVDA VDEQPSENNQ TAADATNEEK 780
781 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
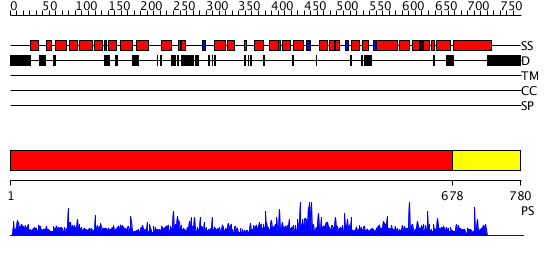
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..677] | 1000.0 | CBP80, 80KDa nuclear cap-binding protein | |
| 2 | View Details | [678..780] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)