













| General Information: |
|
| Name(s) found: |
SPCC1393.05
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 956 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA spindle pole body [IDA cytosol [IDA |
| Biological Process: |
chromatin silencing at centromere
[IMP regulation of histone H3-K9 methylation [IMP RITS complex localization [IMP production of siRNA involved in RNA interference [IDA sphingolipid biosynthetic process [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKVSANNQS GFFKFYIDTA LLLNAEFISS FTNCINSEYN GKVKISYSSS SQQFKVIVYE 60
61 EHKREAFSLF EEIVQKLKRE STVYKKPRLD IHPFGQLHYD FTTLLNFQRY LDLGYGIEDV 120
121 KIKENEDIVK SKVGKKPTSS DFYIPCLIKP KIITPFEWEG NFKQVIYSPT VYNGSFKRLF 180
181 YLPDNNDLLK EFAHATNTKY KASLKDNKIV VLANNDQDIN AMLEKMKHIE NLVSQKVYEY 240
241 PEETLVYAGK IAPCQLKFLK LKFENEILYT AMHTTSIFKS VFNILGSFYT MRLLRNKHDG 300
301 DSYELVPVFA KNPSKTPPCN DNFKIFSDDI KLIGCKIFAG VSLNFSSPKP AHRFYELNKT 360
361 SSNLSIPVLQ KPSNFHSSST ELSDNSIHQG RRAVDPVVNQ NNPSNFEEMI MNKLNKLPTI 420
421 DKQILGTSSL THFQDKTTAI EHSINKSNSK QPPRFKFQLP PRPTSNTLPL EPEEELVTRY 480
481 SVSSDGNTVD EAITKQSQTF QLVNSNEFNE VNANDVHKSL RQNCAKLDFD DSKSKNLLSV 540
541 ECLELDKGSD CSTPKSGSLT PSIDMKFLRL QDEKMDDLGD NYYTILMSSN PVSSYGVGSL 600
601 YLFQPKIVCS EKYINHEEID NMNLKSLHRW LSRSLHVLQS FSGEIELNLE FGVILYPNIS 660
661 SDVSACSHGF MNIYKDLNLP RSYFADCITK SVSNIDSLLN TPVKILFGRT EYTYPLIEHE 720
721 VLDSKNYFVF KGSLLFTDDK RNKTEDSTVF FSMICSSKLD QFAFYKDSKQ SSTCTINFPL 780
781 ALWDSRLRIE TKVPLNDAIL TEFSKSIRFR NVNKDLLLVF GNMEDRVIIH SVTRINENSV 840
841 PFNKNLLPHS LEFILKCSKV KSYDISPSSI NSKEAFVCLD RSKESKPIES CFSLSIQSVY 900
901 MQSQFKYNNS IRAGETAPWK LANKQFIGGA ITENVSDLYT AAIIMVNQLG GIHPTI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
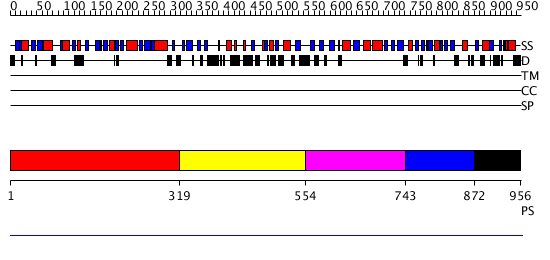
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..318] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [319..553] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [554..742] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [743..871] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [872..956] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)