













| General Information: |
|
| Name(s) found: |
pcm1 /
SPCC330.10
[Sanger Pombe]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 389 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cell division site [IDA DNA-directed RNA polymerase II, holoenzyme [ISS P-TEFb-cap methyltransferase complex [IDA cytosol [IDA |
| Biological Process: |
RNA methylation
[IC mRNA capping [IGI |
| Molecular Function: |
RNA binding
[IEA]
mRNA (guanine-N7-)-methyltransferase activity [IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSHHMMERS LTFERNYSHL FIIIFIIRDM SSSNSRVHEE QPPTENRRYA RPTAQMNRVI 60
61 EQQPRRRDYF QNNDNSGRRG YNRHENNGNA QDVVRSHYNA RPDLGYKKRQ FSPIIQLKRF 120
121 NNWIKSVLIQ KFAPHASDYP ILVLDMGCGK GGDLIKWDKA GIDGYIGIDI AEVSVNQAKK 180
181 RYREMHASFD ALFYAGDCFS SSINELLPPD QRKFDVVSLQ FCMHYAFESE EKVRVLLGNV 240
241 SKCLPRGGVM IGTIPNSDVI VKHIKMLKPG EKEWGNDIYK VRFPESPPRS FRPPYGIQYY 300
301 FYLEDAVTDV PEYVVPFEAF RAVAEGYNLE LIWVKPFLDI LNEEKNSETY GPLMDRMKVV 360
361 DNEGHRGIGG QEKEAAGFYL AFAFEKRGI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
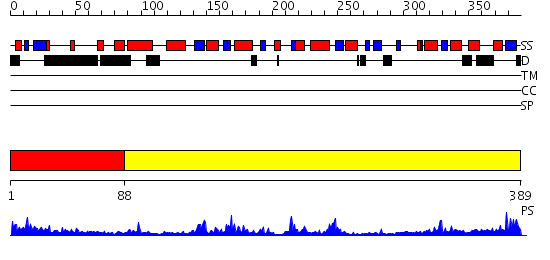
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | 12.69897 | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adensyl-L-methionine (SAM) | |
| 2 | View Details | [88..389] | 36.0 | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.68 |
Source: Reynolds et al. (2008)