













| General Information: |
|
| Name(s) found: |
SPAC1142.04
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 707 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Noc2p-Noc3p complex
[ISS Noc1p-Noc2p complex [ISS] nucleolus [IDA |
| Biological Process: |
ribosomal subunit export from nucleus
[ISS ribosome biogenesis [ISS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKLSSIIKA RKANALAKKN DKIKKKDTDK GIRQKHSKKE GKDETLKRDV EKFIQNVNSE 60
61 DTPSEDDSMS MDAFLEGGFE ELDSANSNDA GSSRKRKNLP NENTQDSTSE SSEEEEDGLE 120
121 SYQKQLEGLK EKDPEFYKFL EQNDQDLLEF NAAETDAMAK EIDENERLKS SSGKIVLTSD 180
181 TIQQWQKLLE TNHSLTTLQK VVQAFKAAAF LNEEEAEDLK YTISDSKVFN DLLLLAIQYV 240
241 PKVLNYHVPI QEDAKGKKFI NTDSKVLPKL RPVLKSYGFS ILRLLEGMTD AKNISLLLRE 300
301 AQNVLPYMIT YRKFLKQFTQ ATVEVWSSTR DDSVRFSAVV LLRTLCLTAD ITLLEFVLKE 360
361 VYLGMARQSA YTTVHTLDKI NFLKNSAVNL FLLDAESCYL IGFRYIRQLA ITLRNTIHQP 420
421 SKDSRKPVQS WSYVHSLDFW ARLLSQAAWL SREKGVASEL QSLVYPLVQI ALGVIMSSPS 480
481 SQLFPMRFHI IRSLIYLSRH TGVFIPLAPS LFEVLDSSYV SRKAKASTLK PLDFDVELRA 540
541 SSSYLRTKVY QDGLIDQLLE LLSEYYVLYA TDISFPELVI PAIVRSKRFA KRSKNAKLNR 600
601 GLLTLVNRLE QQSKFMTEKR NQQKFAPIDS DSVEQFAQTI DWQQTPLGIY VVTQRQTREE 660
661 QRKLIRESVQ QDQEHKEQMR QKKKQALKSD DIELDDLSEE EAEDIDE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
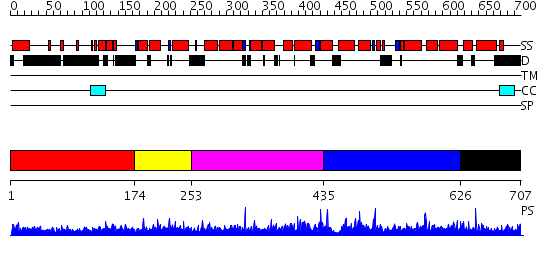
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..173] | 3.154902 | Heavy meromyosin subfragment | |
| 2 | View Details | [174..252] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [253..434] | 1.050946 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [435..625] | 1.050975 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [626..707] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.70 |
Source: Reynolds et al. (2008)