













| General Information: |
|
| Name(s) found: |
SPBC1711.08
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 336 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[ISS cytosol [IDA |
| Biological Process: |
regulation of catalytic activity
[IC]
cellular response to stress [ISS protein folding [ISS |
| Molecular Function: |
heat shock protein binding
[ISS chaperone binding [IEA] ATPase activator activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSATSINPNN WHWTSKDCRV WSHEYFNKEL PKIQASEGPT SARITQVNSC EGDVDVSMRK 60
61 RKVITIFDLK IQMEFKGETK DGVEATGSIT CPELSYDLGY SDYVFDIDIY SASKEKEPIK 120
121 ELVREKIIPQ IRQLFSGFSQ VLLQTHGDDV YLSTEEHNGN AARGLPVHSS FKQNNSSQTS 180
181 SNKGTTTVAA GSGSDGSRVS AVVNTADISE NYTFDAPANE LYATFLDPAR VAAWSRAPPQ 240
241 LDVRPQGAFS LFHGNVVGKF LVLEENKKIV QTWRLSSWPT GHYAEITFTF DQADSYTTLR 300
301 MIMKGVPIGE EEVVQGNIQD YYIRPIKTVF GFGAVL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
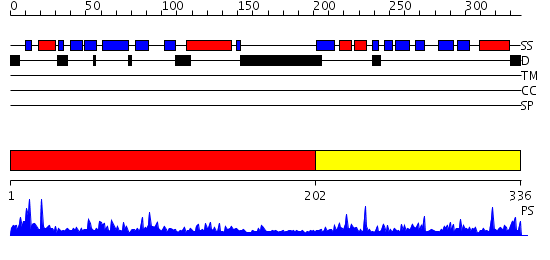
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..201] | 50.045757 | The Structure of the complex between Aha1 and HSP90 | |
| 2 | View Details | [202..336] | 38.09691 | The solution structure of the C-terminal domain of human Activator of 90 kDa heat shock protein ATPase homolog 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)