













| General Information: |
|
| Name(s) found: |
ppk38 /
SPCP1E11.02
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 650 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA actin cortical patch [ISS |
| Biological Process: |
signal transduction
[IC actin filament organization [ISS protein amino acid phosphorylation [ISS actin cortical patch assembly [ISS |
| Molecular Function: |
ATP binding
[ISS protein serine/threonine kinase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNETNNTSLL PVSSLPSGLL PVGFSCTVEK FSVTVKRYLA EGGFSHVYLV QLVFPGKPPF 60
61 EAVLKRIFAT DAMALRAVHE EVRTMKLVSN QKRCVSYYGS EFFRTSKNQF EVLVLLEYCP 120
121 CGGLIDFLNT RLQVRLSEQE ILKIASDVTE AVAVMHYLKP PLIHRDLKIE NVLLAAPNSY 180
181 KLCDFGSACH PIPGAKTAAE AKQLEYDIEK FTTWQYRCPE MINVHKGFGI DEKSDIWALG 240
241 VLFYKLCYYT TPFEHQGLAA IMNVSYAFPT FPPYSDRLKR LISTLLQQYP WQRPNIYQTF 300
301 CEICKMRNVP IHIYDIYNGK NVSSCNPSGS EYLQHASKLE NSGIHQSKSS VFPQPASAMK 360
361 PMASPMLPNV NSMPYLSNGD HNNNGNTSSP VSRFSYGQHT SNVPSTQKLP SNFRVTQGAP 420
421 PSHTYGPPPP VQPKPKISPT TPRLSTLALA DDMFSSTAKE TVPTNEAVFT GDVKSFDSQE 480
481 SNIIESEPLS ASNASGKPRT SVNRLVDRYN HTSSLNKVAA APAPVPKPVN LKSVENPQNN 540
541 ISAPTPSSLQ SSNAPVGLGE VESKSVPPTN MATERGVVGR RASMSIAVNA RRVSKPEKEH 600
601 TNPNAEQGDV IPEKPMSIKE RMNMLMTKTD YEKPKVEGYG RYTDVQQTKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
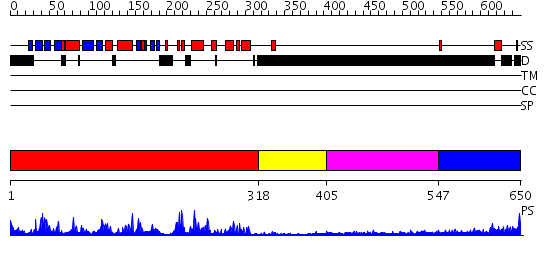
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..317] | 63.0 | Crystal Structure of Aurora-A Protein Kinase | |
| 2 | View Details | [318..404] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [405..546] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [547..650] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.49 |
Source: Reynolds et al. (2008)