













| General Information: |
|
| Name(s) found: |
SPBC106.13
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 404 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA cytosol [IDA GID complex [ISS] |
| Biological Process: |
proteasomal ubiquitin-dependent protein catabolic process
[ISS]
negative regulation of gluconeogenesis [ISS] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNFRPEQQYI LEKPGILLSF EQLRINFKHI LRHLEHESHV INSTLTTLIS QENASMDEKI 60
61 EKIDSLLSRV STVKKKMKHL HDCEALFIKQ TKSRLLFMNR LQGIRDMESA DFLDWSRVRL 120
121 NRLVADYMMA NGYHGAAALL CKDSQLENLV DLGIYKRYQL IHDSILQQEL KEVLSWCSEH 180
181 RAILKKNNST LELEVRLQRF IELIKSKKLC QAIAFAKAHF GTWANEHPAR LQLAAALLAF 240
241 PEFTNGSPYS LLLSDDRWEY LASLFTSNFT AVHNIPSVPL LHALLAAGLS SLKTPLCYLD 300
301 ANDDNPLALQ SQTVKQCPVC TPCLNDLGKA LPYAHITQSA IVDSLTGEGL DSDNCPVAFP 360
361 NGRVYGIQSL ISWNEANGTR EGFLRDPYSG KEFPFQLLRK VYVV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
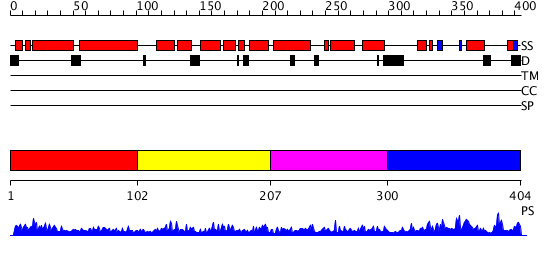
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | 3.098995 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [102..206] | 15.207995 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [207..299] | 5.212996 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [300..404] | 4.0 | Solution Structure of the RING domain of the Tripartite motif protein 32 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)