













| General Information: |
|
| Name(s) found: |
SPBC418.02
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 695 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA NatA complex [ISS cytoplasm [ISS cytosol [IDA |
| Biological Process: |
protein amino acid acetylation
[ISS |
| Molecular Function: |
peptide alpha-N-acetyltransferase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKLSEKEAF LFDRSIDQFE KGQYSKSLKT IQSVLKKKPK HPDSVALLGL NLCKLHDSRS 60
61 ALLKCGYASS IDPKSQFCWH ALAIVYRETK DYNNSLKCYQ NALAISPNNE SLWYDAAYLQ 120
121 AQLGLYQPLF DNWNRLLQLD SSNLEYRLCF TLSAFLSGNY KESLEQIQYL ISSCNLSPLV 180
181 VSRLISFLPR ICEHIENGSQ TVLEILLMNQ NSFLNNFNFE HIKADFAFRQ KNYEESIYLY 240
241 ARLLIKFPNR LDYSEKYLNS LWNFYKSGGL ALDLLLKRTD SLIKTFSEIL QTGISVLIFL 300
301 LSKNLDYDFC LNHLISYSMH HFIPSFISLL KIPLKTNDAF SKKLITMLSN FREGDSAKNI 360
361 PTHKLWCTYC LCLAHYKLGD YEESNYWLNL AIDHTPTYPE LFLAKAKIFL CMGEIEEALC 420
421 SFKRSVELDK SDRALASKYA KYLIRMDRNE EAYIVLSKFS RFRFGGVCNY LAETECVWFL 480
481 VEDGESLLRQ KLYGLALKRF HSIYQIYKKW SFLKFDYFTQ CAEDGEFQEY VELVEWSDNL 540
541 WSSTDYLRAT LGALTIYLLL FESKFNMYGN KAEEISHMSE VEQIAYARED NKKIMKLQKI 600
601 EEDKIKSYIP SESEEPLVID EDYFGHKLLI TDDPLTEAMR FLQPICWHKI KGWGFLKILS 660
661 SKLYKLKGII FCYYALTNNI EGLHQKANSL ETSVL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
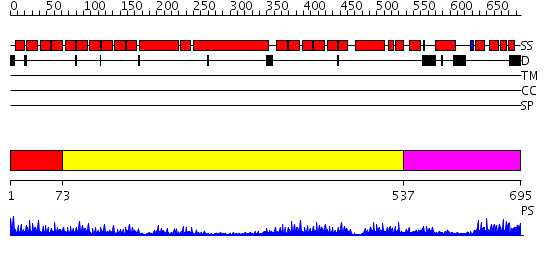
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..72] | 21.69897 | Crystal structure of an 8 repeat consensus TPR superhelix (trigonal crystal form) | |
| 2 | View Details | [73..536] | 33.69897 | No description for 2gw1A was found. | |
| 3 | View Details | [537..695] | 22.69897 | No description for 2j9qA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)