













| General Information: |
|
| Name(s) found: |
SPBC1683.13c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 618 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
regulation of transcription, DNA-dependent
[ISS gene-specific transcription from RNA polymerase II promoter [RCA |
| Molecular Function: |
DNA binding
[ISS transcription factor activity [ISS zinc ion binding [ISS specific RNA polymerase II transcription factor activity [RCA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQMKPRQDKK NQEIFRISCQ RCRQRKIKCD RLHPCFQCVK SNSQCFYPED PIRRRAPKEY 60
61 VEALERQIAF FEAFVKKLAK VGSDEQSLMI QDMNNKIVNE GNEYDQTPDI SARKRRKHFR 120
121 MLPQNNFRYF QFYGTTNVIS ASNLTTTSEI PTFKFPIFSK RKYNDTENLY EQPFHLEFDT 180
181 CQELLSLFFL KQYHNFMFVF RDYFIRDFEL GGGPYYSQWL LFAICSIGAM ISPDDDLKNL 240
241 SNTLANIAEK WVLDEGLNSP DITTLQTLLV LGIREIGRGL TFKGWLFSGM AFRLVYDMGL 300
301 HLDPDHWDHS EESRIDREVR RRCFWGCFTL DKLISLCYGR PPGLYLKQTD VRNTTQLPYI 360
361 SELDEPFEIF NKKSELFAAV SAGEDRRGLV QFWLNQVELC KIIHRMLTEV FEDRTSSVLE 420
421 ASINNIHTEL QKWIADIPME LQWNTRSQKE TSSTVLLLHM LYHSVIIILN RPSDDNYLKL 480
481 DNTERYTFEI CWKSAKTIVQ LLKIYFKKYD ADCLPMTFIH IATSAARIIL VKLNENIPED 540
541 GDVCNIYLEI ITNALDVCAN VWPLASQASR AILNAYKSCA TSPKENNEDS LPLQRSPSLD 600
601 DVSRFDSLDY IFSPNVKY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
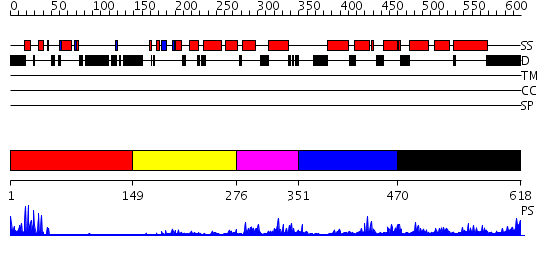
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..148] | 3.09691 | Gal4; CD2-Gal4 | |
| 2 | View Details | [149..275] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [276..350] | 33.920819 | No description for PF04082.9 was found. No confident structure predictions are available. | |
| 4 | View Details | [351..469] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [470..618] | 5.366997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)