













| General Information: |
|
| Name(s) found: |
SPBC1A4.09
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 680 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
tRNA pseudouridine synthesis
[ISS]
snRNA pseudouridine synthesis [ISS] enzyme-directed rRNA pseudouridine synthesis [ISS] |
| Molecular Function: |
RNA binding
[IEA]
pseudouridine synthase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQENHVDVP RKRIRIDQSE SSRNLERNGL EDEANAPSDL SGQKFYMTES DVGIDAFLNP 60
61 NLPSIDGIIK ARFTDFAVFE VDTDGNIVHL TDMEAHDPIL SKATGDKETE DAKDSSNQDI 120
121 SNDQKAPSFK EQEPATLPIL PNDLQSIIPK GIGNEFIQSL HKLSVGEITD PISLILPENT 180
181 PPMDKGQRTI LHQFIRNNFS GLESSTKGNG TFTVSKTTRK NQPRSRRDPR LSWKALGGEY 240
241 CHFHLYKENR DSMDCLGKIA RLLKVPTRTL SIAGTKDRRG VTCQRVAIHH VRASRLAQLN 300
301 SGSLKNSTYG FLLGNYSYKN SNLRLGDLKG NEFHIVVRNV ITPKEKVVEA LNSLKEHGFI 360
361 NYFGLQRFGT SSVGTHTIGV RLLQSDWKGA VDLILSPRPE HTGSVKEAID LWHSTHDAEA 420
421 SLRILPRRMI AESSILETWS RSGNQTDYLG AFQRIPRHLR SIYPHAYQSY VWNRVASWRI 480
481 KNLGDRPVVG DLVYSTESNG LSQKSPIVDP EAPDLLEDLP VSSKLSARPI EEDEVNNFSI 540
541 YDIVLPLPGR NVIYPKNETF DIYKSVMNEA SLDPLNMSRK DRELSLPGDY RKLLVRPENM 600
601 EFNFIKYDNM EQQLILTDKD RLENRSISVS SEVGKHTAVT LKFVLPSSAY ATMALREALR 660
661 TATASGDQRM LMPAVLKDSI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
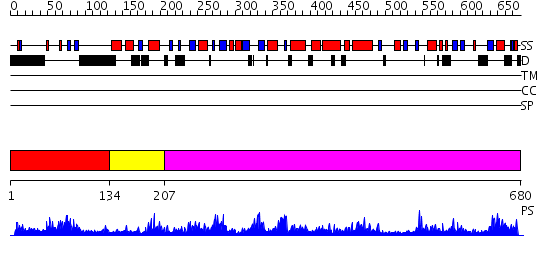
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | 2.049996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [134..206] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [207..680] | 102.0 | Crystal Structure of the Putative tRNA pseudouridine synthase D (TruD) from Methanosarcina mazei, Northeast Structural Genomics Target MaR1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)