













| General Information: |
|
| Name(s) found: |
tea3 /
SPAC6G10.02c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1125 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cytoplasm [IDA cell cortex of cell tip [IDA |
| Biological Process: |
selection of site for barrier septum formation
[IMP establishment or maintenance of cell polarity [IMP activation of bipolar cell growth [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQKVLSRQS DNSQDVSAEQ LDVVESGSID QQNIRAWVVR KVKENDKRTS TNQSFKWEAV 60
61 KPASCLDAAN EKFMYLHGGR EKSGISNSLF KLDLDSCTVY SHNRGEDNDS PARVGHSIVC 120
121 SADTIYLFGG CDSETDSTFE VGDNSLYAYN FKSNQWNLVS TQSPLPSPRT GHSMLLVDSK 180
181 LWIFGGECQG KYLNDIHLFD TKGVDRRTQS ELKQKANANN VEKANMEFDE TDWSWETPFL 240
241 HSSSPPPRSN HSVTLVQGKI FVHGGHNDTG PLSDLWLFDL ETLSWTEVRS IGRFPGPREG 300
301 HQATTIDDTV YIYGGRDNKG LILNELWAFN YSQQRWSLVS NPIPILLSDS SSYKIVSKNN 360
361 HILLLYLNAL DAPKQLLCYE ADPKNLYWDK DKFSDIPVLQ HISMKPSNAS NHTVSLGYLN 420
421 DRPNHSKKNS VTSTSSSQFN NFLEQNQKAV RSARHRHYAS LDEQGLHSLR NLSKTSGMNH 480
481 SADFSLHEFG QADPFAYEIE KPIASLPLPN GNDTISRSSE SSSPINESES NSLLKLQSDF 540
541 KFSNSDDRVA WLEEQLLYCM QQGYTLKPPN LFQHVDEKLR LEKKEQLSYL EILKVIEQML 600
601 ESNEQKFKKQ IVSLASENAK LAAQRDAAVE NANYSRSLIQ KKTTDETVGS LIEKVGKLEY 660
661 EVQGTLEEAT SYYQKNTELQ QLLKQNESAS ELLKSRNEKL CVDYDKLRSV FEEDSSKILS 720
721 LQKENENLQS QILQISEELV DYRSRCEALE YGNYELETKL IEMHDRVEMQ TNVIEASASA 780
781 LDVSNTAILS FEDSLRRERD EKSTLQQKCL NLQYEYENVR IELENLQSRA LELESALEQS 840
841 VSDAKYSKAI MQSGLSKLLS SINENKDNLK EFSKSKQKIS YLESQLEGLH ELLRESQRLC 900
901 EGRTKELLNS QQKLYDLKHS YSSVMTEKSK LSDQVNDLTE QAKITQRKLS EVQIALADSK 960
961 MNQQLSGKDS TDVHLPTDFS ASSSPLRSYF NEEDSFNNAS AAHSSKESDI PSGGVFTKYR1020
1021 NHFGNLMTSE ETKAPDNNDL HKRLSDVINS QQKFLSLSPQ VSKDYYDVRS KLNDTAGSFS1080
1081 GEEMRAIDDN YYASRIKQLE DDYQKAITYA NCSDESFQQL SHSFM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
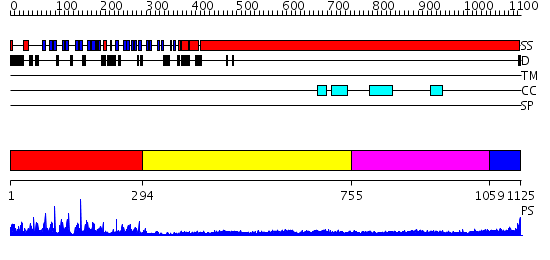
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..293] | 51.0 | Crystal structure of the Kelch domain of human Keap1 | |
| 2 | View Details | [294..754] | 1.23 | COLICIN IA | |
| 3 | View Details | [755..1058] | 18.045757 | Heavy meromyosin subfragment | |
| 4 | View Details | [1059..1125] | 14.09691 | Tropomyosin |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)