













| General Information: |
|
| Name(s) found: |
SPBC582.10c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 830 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA cytosol [IDA nucleotide-excision repair factor 4 complex [ISS |
| Biological Process: |
nucleotide-excision repair
[ISS |
| Molecular Function: |
DNA binding
[IEA]
ATP binding [IEA] DNA-dependent ATPase activity [ISS helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKASKNKES GVKKANNSFL QNFGVNTAGK ENESTSLPRL PKSEDESIPK QSIKSKNKKK 60
61 TQHFLSDAFE SLKRQEIDTN EKEVIVSAPI TSKPAQVFYS KKLERKDEGI RKWEKDPFAP 120
121 ISVKSGAWKK PYTKVPEVSI NKATSKRTDL INDKPIVIPI PRASTSTLYF GKHNKPTSEN 180
181 RKGPIGIPTE EILTSQNTQA MLHKLFENNV LDNVKDDSMQ RQSSFIPGMH IRLLDHQVQG 240
241 LTWLKSRETV SKSSASGGIL ADDMGLGKTI QMIALILSHP LPKKKHSIKS TLVVAPLSLI 300
301 KQWESEVQTK SKLTAIVYHG ASRYKLLKVI HEYDVVITTY QILVSEWVSH NTTGTDGKSP 360
361 TEAKSYEKKK PSLFAFYWWR IILDEAHTIK NKSSKSALAC CALQGINRWC LTGTPLQNNV 420
421 DELYSLVKFL HINPFNDQSV WKDQISLPLC QGEENLVFKR LRMLLSVIML RRTKTLLEAN 480
481 AGKDGTGGAL KLSKRLVYKV ICKFEESERD FYSNLARNME RTMSNFVNSG KLGKNYTNIL 540
541 CLLLRLRQAC NHPQSLNFQF EQDVDAFNAL DGAANTNKLA SDQDVDDLAN LLETVEIGSR 600
601 KKSFCTICMA ELPPDFHEKK CKDCSRNFKE LDKGIQDPND KTLYKSSKIR EILKILSLDE 660
661 QEEDDTVRGL RKTIIFSQFT TFLDIIDLHL RKAGIGFVRY DGRMNNRARE KSLDLLRSDS 720
721 GTQVLLCSLK CGALGLNLTC ASRVILCDVW WNPAIEEQAI DRVHRIGQRR DVLVYKLVVE 780
781 NTIEEKIVEL QNLKRDLAKQ ALGDGKKSVF TSKKLTLNDL LFLFNKRAAA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
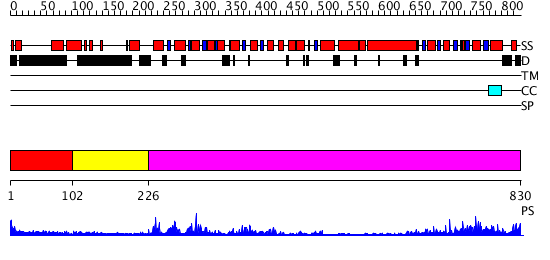
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | 1.344993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [102..225] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [226..830] | 41.045757 | Nucleotide excision repair enzyme UvrB |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)