













| General Information: |
|
| Name(s) found: |
SPAC26A3.10
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 923 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cell tip [IDA cytosol [IDA |
| Biological Process: |
small GTPase mediated signal transduction
[NAS]
regulation of GTPase activity [ISS regulation of ARF GTPase activity [IEA] intracellular protein transport [RCA |
| Molecular Function: |
zinc ion binding
[IEA]
ARF GTPase activator activity [IEA] GTPase activator activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGSDSLLKS VDVSKLDNGS TISFRAREDG FLESISSSSG KLVDMVLDST SKECLPKFSF 60
61 QNKLDFYFLF SDRVLQLQKE RKVLVLIHCE SLQSFDSFFA QKLKYADLIS SDNVHEIFLP 120
121 DSNAANIRYH WEWSPPSGKL LEYECKNYSC IAVYSTEDCT LERISKFSYY TVSRCGDSMD 180
181 DTFFSESQRV TSPLTTSQTV QTQPPQSPEA KTELSLINTK TVIFPENYED GPSFRSMLHE 240
241 LEQKSSLMKY YCKKIMKRIV QLSDAYDASQ VAVMKLSETL SEASNSTSMN MDILLDSYLT 300
301 KAMDIHATFI QKLNYDLINL LYEPFHNIYS SFIKPIDDRR LEFDEQSKSF YGSLSRYLSA 360
361 KKDKKGGDSK FFQKEKTFAL QRYDYYCFMQ DLHDGSIIND INGIFLQYFH RQYDHIALFS 420
421 NLMNSVLPNL QQLNLKLEKT KWSTTRRDKG REMHRSQVIQ TSGRPKSMAP PSPSPISPSF 480
481 PLHEIQSPMP NRRMAASADD ISQTSNFTTE IKGKCISNGG SASPDKIFKE GLLLVFGATE 540
541 LGTDLAMVSK AAWHKHWIVV ENGSLWEYAN WKDSVKSNVS SISLKHASAD KVRKQGRRFC 600
601 FEVVTPKLKR LYQATSAEEM DSWIEAICEA AKISSFQLSR VATPLSASVR RPSKVFPLFS 660
661 TSFETTPISR KLSGSGIKKA FSRKGSWNLQ QFFRSDNSGT MHMEQLERYH ASANIFIQML 720
721 RKTDVSNSVC ADCGSVKDVT WCSINIPVVL CIECSGIHRS LGTHISKTRS LLLDSLSQQS 780
781 KVLLCKIGNA AVNRVYEKGL SNPSLKPKPE HNAQVKLAFA QKKYVEHAFI DFAGVDADAT 840
841 LLEGLEQNKI SKILLGLAAK PNFEENGVVF LKAVTRDTSK LHLLELLFMN GLLLPDSEQL 900
901 SEHVSPDMQS YLSQKQFTKY LKE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
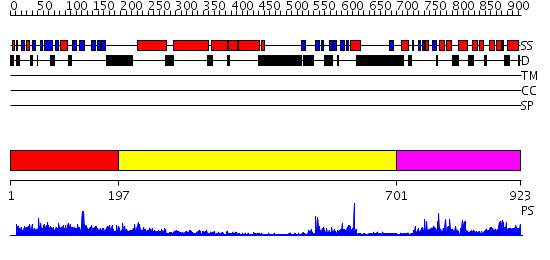
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..196] | 2.065952 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [197..700] | 30.0 | No description for 2elbA was found. | |
| 3 | View Details | [701..923] | 44.69897 | Crystal structure of UPLC1 GAP domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)