













| General Information: |
|
| Name(s) found: |
SPAC11D3.07c
[Sanger Pombe]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 603 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQNKACDLC RLKKIKCSRG QPRCQTCTLF QADCHYSNRA RRKRLVQRSK ETFGGITLPV 60
61 KNIEKPEDGE ESVQRRLKLL EEKIDLLLDI ATETSEFKRE NKAIELPSLV TQIKDAESIV 120
121 IKHRQGSPVD NTPTRILNVE SLFPPQLPDW EKAFHDIPKK EVAHELVTSY FQHVNWWWPT 180
181 FVYNDFMYEF ERLYAFGFHS NNAWLISFYS ILALSSIRKR LGNSKTLAES LFSTAWVFVQ 240
241 KSDFFLTPSI DKVQALIVMT QYAAYLSSSS LCRTLCGQAC LMAQQLNLHR KQSTDVEPEK 300
301 AESWKRIFWM CYILDKNISL IFGTPSVFND KDIDCNLPDS KYELLFGVQS GGDLIFVPTV 360
361 SLTIIQSEIR NRLYSVKSPT QMAAREKIII PIHQKLKAWE ENLPSEIKMY HEMLLNNTFS 420
421 PTISLSDRFE FLTFAGMEVY FSYLNTLIIL HRPSSSTENR RICINAAREA VQLLKNRLNI 480
481 DLRVNVKADP LWIFLYCPFT PFLIIFNNLV HETDTETDSE TLLNDLDLLH VIYDFFMEME 540
541 PVSDVALELV KIADKLLRVA KEVCSAKNND VTDSTFKDIV EGFELNDLNS WDFDRVTNVM 600
601 RNL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
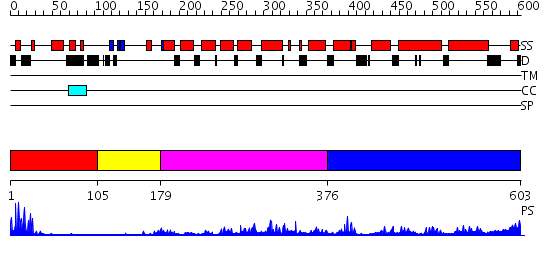
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..104] | 8.39794 | PPR1 | |
| 2 | View Details | [105..178] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [179..375] | 34.013228 | No description for PF04082.9 was found. No confident structure predictions are available. | |
| 4 | View Details | [376..603] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)