













| General Information: |
|
| Name(s) found: |
SPAC2G11.10c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 401 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA cytosol [IDA |
| Biological Process: |
cellular response to oxidative stress
[ISS protein urmylation [IC |
| Molecular Function: |
ATP binding
[IEA]
URM1 activating enzyme activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNPQEKIICS SNGLELSLDE YSRYGRQMLL SEIGLPGQLS LKRSSVLVIG AGGLGCPAMQ 60
61 YLVAAGIGTL GIMDGDVVDK SNLHRQIIHS TSKQGMHKAI SAKQFLEDLN PNVIINTYLE 120
121 FASASNLFSI IEQYDVVLDC TDNQYTRYLI SDTCVLLGRP LVSASALKLE GQLCIYNYCN 180
181 GPCYRCMFPN PTPVVASCAK SGILGPVVGT MGTMQALETV KLILHINGIK KDQFDPYMLL 240
241 FHAFKVPQWK HIRIRPRQQS CKACGPNKML SREFMESSPK EYTTICDYVP TLSKQLAPIR 300
301 RISALDLKNL IETSPHITFL DVREPVQFGI CRLPLFKNIP LSEVDSLQNL SGKVCVICRS 360
361 GNTSQTAVRK LQELNPQADI FDVVAGLKGW STEVDPNFPL Y |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
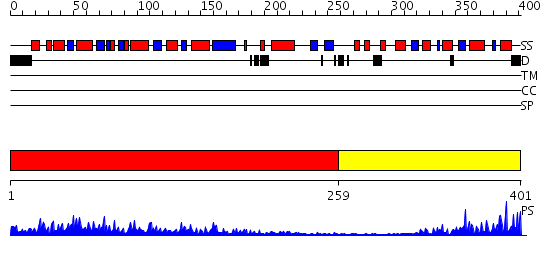
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..258] | 62.221849 | Molybdenum cofactor biosynthesis protein MoeB | |
| 2 | View Details | [259..401] | 15.154902 | Crystal structure of Rhodanese from Thermus thermophilus HB8 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.67 |
Source: Reynolds et al. (2008)