













| General Information: |
|
| Name(s) found: |
gef2 /
SPAC31A2.16
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1101 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA spindle pole body [IDA cytosol [IDA |
| Biological Process: |
regulation of Rho protein signal transduction
[IEA]
regulation of catalytic activity [IC |
| Molecular Function: |
Rho guanyl-nucleotide exchange factor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDVGTMGSRA ELLAISIQFG GLWVFRDLLQ MNPLFEQDHV FCTFNHPNMG LVLISFAKVD 60
61 GHILAAYALS KLGPDTFSIC YPFSSKLDAF AQTLDLEWQI ASMHKEAAAC VYEFLCVESV 120
121 CLHTGENWRE SVIEPEYASY IFDTINVFQS SSLTRELYSL GEPNGVREFV VDAIFDIKVD 180
181 NSWWEDPSNS YWKTVIGSRE MFEDSRKKTS SPSPSFASSK DAGTIPAIQK KKSLLIEMME 240
241 TESTYVERLR HLVNDYALPL RDLAKSSKKL LGLYELNTLF PPCLNKLIQL NSAFLDEFEA 300
301 IMSDLNFEDI DEKKFEEIDL RLACCFESHF FAFSQHYPRY LEQSNDFGNV LKMASKIPKF 360
361 VEFHDQVKLN ANMNVGLSQL IMEPVQRIPR YSLFLDQIIL LTQEGECQHT YVRSVEIIKN 420
421 IAEMPTVDAE ERSRIFAGLQ HIIPDLAPNM ISNSRNFIDC IDVTREFLKN GQLHLIPYTL 480
481 ILFNDRICLV QRRSKSSIAS TILDLRKQNP RNSYSKEKRA QYIGSNMNEA VELTRSMVEE 540
541 NTIFLISKYA SSPSFFNEYP ILKFRCDFEN VRTMDRFYQS FQKALSMNKS QPSCLSFSKL 600
601 NDFVVFFNNY SRFEYEKESK RSDIVCICTN DANVDKHKFL QDGNIVITFF QQDEDFHLSF 660
661 DSWLGVSLPT EAVIAKEDLR EACLNYLINI KRLLLCPFSN RNFSSLDLYS NLIQHLLSAN 720
721 SSPRKSRLSF GGRPGSPSKI SLSLNRFYNQ GGLSKSCATL PSQMYNLDHN NISQKSLKFN 780
781 THNTSKASAE KTVEHLEAFK GGFKYHTDLK NLLYPLSEKE KIEGDELYDN ILKETFNEEL 840
841 LSHYPPNIIY ATFQKYLSSF INRKFGVLLS SSFIQQLNTV ENLNLSFNST DAVYHLKKIL 900
901 QDLPESSLKI LENIFSIASD LLLRLPLKDQ CDFVTKQLAI ALAPSMFGSN AVELVYYLAY 960
961 HSDRIFGTVE ELPTPVSPAN SNNDKQLDES KFQAIAMKEM PERHPKEILP GQIEREAYED1020
1021 LRRKYHLTLA RLAQMTRLNE DSKKSIPLLY DRFNHDLKLI KQSVQASLIR KQCELDTAKW1080
1081 TLEEYESKLN AKEGCQTNIF I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
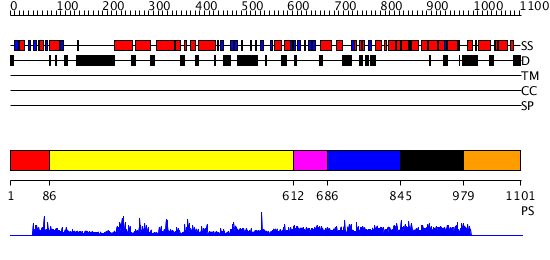
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [86..611] | 63.0 | No description for 2pz1A was found. | |
| 3 | View Details | [612..685] | 5.0 | Solution structure of rat Kalirin N-terminal SH3 domain | |
| 4 | View Details | [686..844] | 1.038968 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [845..978] | N/A | Confident ab initio structure predictions are available. | |
| 6 | View Details | [979..1101] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)