













| General Information: |
|
| Name(s) found: |
ubp21 /
SPBC713.02c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1129 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
regulation of protein kinase activity
[IGI cellular response to stress [IEP ubiquitin-dependent protein catabolic process [ISS |
| Molecular Function: |
ubiquitin thiolesterase activity
[ISS cysteine-type peptidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLSNVDAEE VNMDSSMELE ESSQEPLRAD NYEEIYNSLV HHEPDLEEAA HASYSWVVKN 60
61 FSTLEDKTYS PLFKAGHTTW RIVLFPKGCN QTEYASVFLE YLPQCKVEAI RKYEAELAAG 120
121 KTPTIDPEIV NDETYSCCAQ FALSLSNVQD PTVMQINTSH HRFRSEVKDW GFTRFVDLRK 180
181 IAVPTPEFPV PFLENDEICI SVTVRVLQDP TGVLWHSFVN YNSKKETGYV GLKNQGATCY 240
241 MNSLLQSLFF TNIFRKTVYK IPTDNDDSRD SVAYALQRVF YNLEKQREPV STTELTRSFG 300
301 WNSFDSFMQH DIQEFNRVLQ DNLEKKMKGT EVENALNDIF VGKMKSYVKC IDVNYESSRV 360
361 EDFWDIQLNV KGMDTLEDSF RDAIQVETLT GDNKYYAEGH GLQDAHKGII FESLPNVLQL 420
421 QLKRFDYDML RDMMVKINDR HEFPLEIDLE PYLSETADKS ESHVYVLHGV LVHGGDLHGG 480
481 HYYALIKPEK DSNWFKFDDD RVTRATIKEV LEDNYGGEPA GRAKGYNGNP FKRFMNAYML 540
541 VYFRKSRLDH ILSPVTAEDV PFHVRNTLDE EHRVVERKLL EREEQQIYRR VRVLTTDGFK 600
601 KYHGFDMTDF SASDDDPVLI TTKIKRNANI WDLQKHLAGL LNRDTSGIRI WLMTNRQNRT 660
661 VRVDLPLDKK TILVDQICDM HIRKDMDMRV YVEFLSEHNQ LLADFGATDD NDFDTYIFLK 720
721 IFDYETQQIS GLADLHVSKN SPISSLSEWI REHLKWSSDV PITYYEEIKT GMVDVLDPNA 780
781 SFEKSEIQVG DIICFEKKLV HDSSSDTSHP YKSALDLYDF MAHRVVITFE PRYSDDTNNG 840
841 VFDLVLTTHT NYTDMARAVA NKLNVDPNYL QFTMAHLPSR TPRSVIRNPS KFTLQNAIPS 900
901 TYSHNQNVVM FYEVLDITLS ELERKQLIRV HFLSNGISHE TQMEFYVDKE GTVEDILRQV 960
961 TQKVPLNAED ASRLRLYEVY NHRILKSHLP TDGIYDLNEF STAYVEVTPK EEQMQLKTDD1020
1021 AVSIVVQHFF KDLSRLHDIP FYFVLLRGET LKDLKKRLQK RLGYNDTQFS KVKLAVLQAQ1080
1081 SFGKPYYLTD DDEVLYGELE PQSHILGLDH PPANGSAQYH GMDQAIRMK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
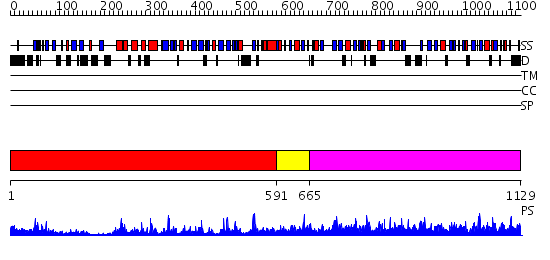
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..590] | 92.69897 | Crystal structure of HAUSP | |
| 2 | View Details | [591..664] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [665..1129] | 2.39 | No description for 2ibiA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)