













| General Information: |
|
| Name(s) found: |
spo3 /
SPAC607.10
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1028 amino acids |
Gene Ontology: |
|
| Cellular Component: |
prospore membrane
[IDA extrinsic to membrane [IEA] |
| Biological Process: |
ascospore formation
[IMP ascospore-type prospore formation [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGILSVIRIF IYCLIRLFSF NSRKRNSSID ELERGEINAF ACDKQNTPSP NSECRPLTPL 60
61 NSPFRRTVEG DTANLQAPSP TACPSFDSAS SIKSSKEDIA CRKLSTERFF YSPNGIDEMK 120
121 KQQQFKKIPT VSITEVEPAF IPSNMEQALF SEEPFVIQDD LSHQINLDDN RKFSRIQKNK 180
181 LDPLITVNLV PIVNGENCFN FYETQMETNF LSPEFIKKPS MVDHKRDSEI NAVTAVSADG 240
241 SFSPLTETLS STISTLSNDS LDGLSSQKNE FTGFNSNSEW IPNDTVDYIC NSDASSVVSN 300
301 LSDFEDCFDQ FGVYDKEYEK KIEKTKRNFP KFQSSATYPP FSAHHQARRN NPQGFRNVKK 360
361 RFQLFKDDCT ALTDSDFLDA LPFFNARVII ALYVEWRLRV YETMIIKGEE SLKNLQRARD 420
421 GPHNKLWLGV FGNKDTIKKL SNYDRRAISN PYVSNHLNFN VRRVKSDTVY IPTILQLLRS 480
481 STQLMTEQAF VASSNLKSTK GLRETRLIQK YKVKGDFEYV YAALYYAAFT QESTLKAVTI 540
541 DDILSEDELE DWWYLNLRYT SFITPQLCFE FLDKEADSCR DRLTEVKSEP PKAVIYEEPL 600
601 NKLSRFSSDR LFTSHELVAI AELISLNEPP LESGKKFYYE EFQKACQKKR DDYRYSPEIE 660
661 FKAAFREKFL QSNEKVPFPR PGEIYEKRCD YFSKYACQNI FISKPLKRSG SIYEVFSDGI 720
721 GRVIYGSVLD YEATRNNILL HFGFEIHCAP SEEELIEREE AFKDFHNMLS FKYSANEIYE 780
781 FCGTHSRAEV HKNEVLKRMA YYLIDENKEI SILRRILSLR IVEPTTSFTR YEYDLWRTFY 840
841 PDVLYLNYSK SGRIPTGSPY IMNGQWRKDL CVSTPPMVDI NSALFPHWFK LSASNLFAGA 900
901 EHAGLNRQKP DKIDLDLYEP LPENSYLVAA ELRVLRALRH KNPENIPLLE KWEVRALHQL 960
961 MAKIRKYYTV PTDYLSLRMD GLQAMLRKRE YMSCYTFNYF DYACLMDHAY RLYEGFNNQV1020
1021 VNLPPRIM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
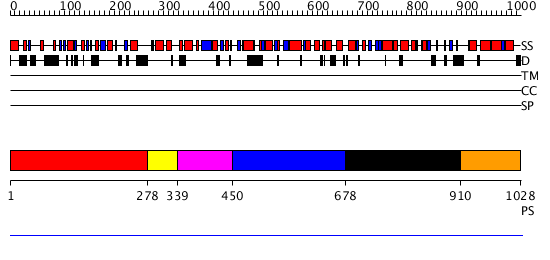
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..277] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [278..338] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [339..449] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [450..677] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [678..909] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [910..1028] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)