













| General Information: |
|
| Name(s) found: |
sec8 /
SPCC970.09
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1088 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cytoplasm [IDA cell cortex of cell tip [IDA cytosol [IDA exocyst [IDA |
| Biological Process: |
vesicle docking during exocytosis
[ISS cytokinetic cell separation [IMP intracellular protein transport [IC endocytosis [IMP exocytosis [IC][IMP |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTRGYSETK KGRYPVGKKS LESPNGYSNY GTSMDNELSS EYASRGMHIG DLENLVNEIE 60
61 DEWKDLGRED YQPISTALEL LDDSSFGRDY KSFLNVYDRI SAALQTIAHT HKDDFTRGIS 120
121 AYGEIMEGIQ KCNSRIIALK QSLEASQECI GNTNSKELQQ TLARSSQYKK VISVLKELNE 180
181 ANQLFDNFHT LVDSKQYYHA SDLIRRVWDE LSRSDFDGIL VVEQFKSRMT GLLSHLEDIL 240
241 SEELVSITFL KDAVAYPIVS YCSPNPLRET SNPYFLRDFL KNNANTSTLG QSEQLRYLEE 300
301 ALSLKLSDCL KMDYGRDSLR DIRIVLESLN LLGKLPNAIS SLKSRTSAEM FTTVDSTSRA 360
361 IVNKYSLGNN VSTVNPFSKS LYDIGLHAET DREHTMISEF LTNLFTKLRC VLMHYRGISE 420
421 FMTKLETKTP KHASSSHKSS IMSVNSDPTS PKVSKFDTSD STFPFDTLLQ AFESEIRLML 480
481 KDYLISKEEY IENSGNFVVG TEMSIYNLPG ENEEDKLFDV TNEIAVENKS NAFYARINEL 540
541 VNEKAPELIL NKSNASVSTI ELFSGSSKEI VRLAGHVVFV GPSVFHASSV LPQTVFFLED 600
601 SVSILKNPNI PPQFAVNFMK EFLRGSYIPQ LYKFMSSHFD TIMKDVGAFQ LHRDWKIYSK 660
661 IPIFKCHVAI VQYFHDLQDY LPIVALNLVE FYELLHTLLV RFRNHCSDYL SDLCRTAVLK 720
721 EYKHVNEDTE DVDDTVRVKL LHDDVTYPQF IKFLKQKNPS LEGLNELCRM ENKRLLQYED 780
781 RAITSEVKLP VSVLSKDSDL VNSVSYLHNS MEWFLQRCFS RFMNGSRRMN VLQQNQANFG 840
841 GDFLPIDNLL GNNSDLMKGY VTVCIELYTD FYKSAYKEVF DSLQRLQFDA LLLIRMEVRL 900
901 QYIHSINQSV NLPDYVVEYR GRPDASIMAL NSTIVTTNLK LETCLNEWER RFVFQGLSEL 960
961 VDSSLYSIFY KIESMNRGSC LQMLKNMSAM IQILKTVKEI HGDVEFPKSS RVFGIYQNGA1020
1021 KKIIEHFIAA PKKELLPDVK QMVRIYYQRL MKDAKRNGRD DLYRQYQKKI GSVLTQFDNT1080
1081 VGGARKNP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
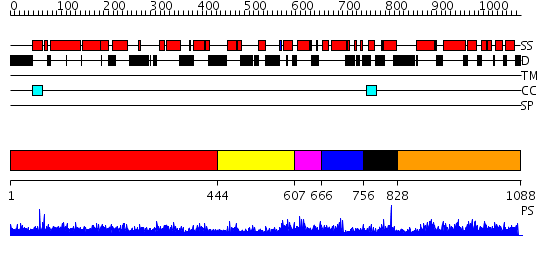
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..443] | 1.03 | The modeled structure of fibritin (gpwac) of bacteriophage T4 based on cryo-EM reconstruction of the extended tail of bacteriophage T4 | |
| 2 | View Details | [444..606] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [607..665] | 1.041984 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [666..755] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [756..827] | 1.045992 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [828..1088] | 1.043979 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)