













| General Information: |
|
| Name(s) found: |
scw1 /
SPCC16C4.07
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 561 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
regulation of barrier septum formation
[IGI regulation of fungal-type cell wall biogenesis [IMP |
| Molecular Function: |
RNA binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFVGSPSAIE KPLSLSSKTK VGDETEFTED ALRGSSLVPT GILNGDDECH ALHMSPQAIH 60
61 SQAGKAESDG LPTYASVEKS TTPIGRLLSN LNGGLASAPP KVGFGFPRYY ALRIEDLPRD 120
121 LTPREFLCTF LFATNVVSVE LNPVSVDNEV AHGLAVFSSR DAAASARDTL LSSDVYSACS 180
181 MIILDNYRKG SQTNLSDETE GSESSVSFNR LSRNHSPTRP LLGNRDLFRR SSNHVSASMP 240
241 TANHSESAYR HSKTPLGDST FQAPNNGGSL HSDRLWSSFP VSYPLTLANV LAKDEVGSPT 300
301 WSPTPSKSST NLRQDGVPPI LRFNSLSINT NVARNYLSSE KGYSAHTQNS SAQSPHPRVF 360
361 SANSAFSTTS PPPLTPSTSR DYPFSASTIS PSTPFSAYSS SHGIHQRIPA STPTNTNPAD 420
421 QNPPCNTIYV GNLPPSTSEE ELKVLFSTQV GYKRLCFRTK GNGPMCFVEF ENIPYAMEAL 480
481 KNLQGVCLSS SIKGGIRLSF SKNPLGVRSS SSSHNNHNGN VRNLHSGSMN NYNTDSLLNH 540
541 TGGHNEVHAS PSWGNNLMYG K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
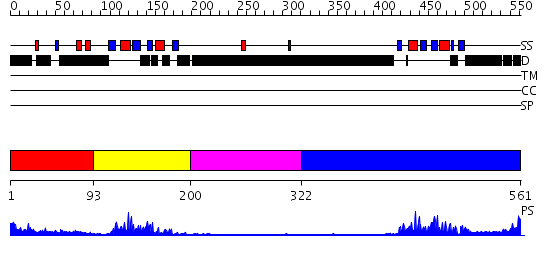
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..92] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [93..199] | 13.522879 | No description for 2do0A was found. | |
| 3 | View Details | [200..321] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [322..561] | 17.154902 | Sex-lethal protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)