













| General Information: |
|
| Name(s) found: |
rst2 /
SPAC6F12.02
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 567 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA cytosol [IDA |
| Biological Process: |
cellular response to nutrient
[IDA induction of conjugation upon nitrogen starvation [IDA regulation of gluconeogenesis [IDA gene-specific transcription from RNA polymerase II promoter [RCA positive regulation of transcription, meiotic [IDA |
| Molecular Function: |
transcription factor activity
[IDA zinc ion binding [IEA] protein binding [IPI specific RNA polymerase II transcription factor activity [RCA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTRESLAPIA SKANTLSESK VSENLMSINS DSGTSNANTP SSVTSNSKPV ASSTAAKKDP 60
61 NAPPQKVKQY VCETCTRAFA RLEHLKRHIR SHTNEKPFTC SEIDGLPTGC GRQFSRRDLL 120
121 LRHQQKIHRN PQPRRRRRST TALPNPSLSN VSVSTTNLAS KPVISLPQAD SIDKFRYPKL 180
181 HAALQAQLAN NSGSFSASWL QAQQQRLVSA GNGRETVNAS ASGAVNPTSS QWSDQRLGVP 240
241 YGPDSPLYYR RATIASDLRP SVYQHPQLYD FNRPESLSGQ LDDESLARMP YSPNAVSRNY 300
301 AMNMTLPESI PEGYEIDKLD WMSFSESLNL PTFNQPSGPS DVSASFLNLQ AMPSPHASPK 360
361 DSLINKSNSY FFQNPPKSNS VDYTRLDNLE HMRQSCISPS GTNFSPSCYS PESLMQQPLT 420
421 LSPSSIPSGL PTYAHVNSPL GTEQYPSEAY PPKGYVDMEG RISEGIKEEG ISLSEEITDK 480
481 DQAFASMGYF DSYNFEGVEN SNSLNNEGDQ METNFQSEKL DNSNSIPFFN FNSFNNNGAN 540
541 WSSDFNYPVS ETSGLLTDNN RNQPPSF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
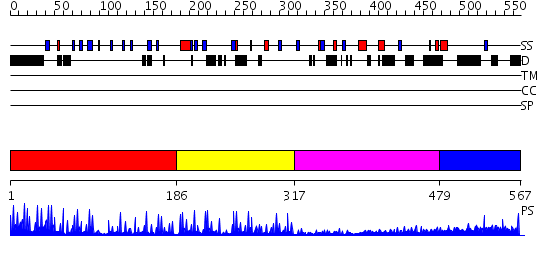
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..185] | 32.30103 | No description for 2i13A was found. | |
| 2 | View Details | [186..316] | 9.522879 | No description for 2yt9A was found. | |
| 3 | View Details | [317..478] | 1.311993 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [479..567] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)