













| General Information: |
|
| Name(s) found: |
rap1 /
SPBC1778.02
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 693 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromosome, telomeric region
[IDA nucleus [IDA nuclear telomeric heterochromatin [ISS telosome [ISS |
| Biological Process: |
meiotic telomere clustering
[IMP protection from non-homologous end joining at telomere [IGI telomeric 3' overhang formation [IGI telomere maintenance [IGI transcription from RNA polymerase II promoter [ISS] telomere capping [IC chromatin silencing at telomere [IMP |
| Molecular Function: |
transcription factor activity
[ISS]
protein binding [IPI specific RNA polymerase II transcription factor activity [IC] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFTFTKSDG SSILFAVSKN FEHIRGFKNA IDCFKGKIEF LSFDKVDPTK HDYYLVAEDE 60
61 RVSDLDIPKG FFERNPEFRH KMLKIAWITQ CIEQGKLLPT ESFEVELNQD DVNRTHDGFR 120
121 KRELFTLEDE KILIDHVHKN DINRFGTKVY EELARKYPQH SLESWRQHYK YMKKRLPPVS 180
181 DSDESNYCQR IIVKPYSSQK DYTQSTHEQT LSSPISKSAS VSKSENKALV NNKRYSDSYF 240
241 YFSKMRRISI DVDYVDEDLN LINAYLSQFG KKRSLNELCA LLSRRFSNRH TFSEWRALFM 300
301 HFFPFINSEG VDPAILSDRE TSAMLDETSD NEVADIDDQM IERKYLFSAS EPNTVKSTNR 360
361 LIFSERKAYA ADDSIDNTPS KVPIVNSLSD PRTNRPFFYS NPDSMYRSIS NPLHLVDSQH 420
421 LSPLNRKTHF NNPIGQPQFT CLDDHEKTLR ETSFRSLDDM SLRKSNSDNI FVKPGEDLEI 480
481 PLLSDYSDSE NISEKSSDDE EAFEKQVTSS YSSPIKVKSQ GKSSKGSSGL DVREHEGSDD 540
541 DAEVFVDRSP ESFGATKVAH TSLEGNAASH KKVEENMQQP VTKKQKKYRM VNEEAHTGPT 600
601 IIIPSDNNEK VTTLPAGHVP SEEKGKFINL AMHELQNEVS ILRSSVNHRE VDEAIDNILR 660
661 YTNSTEQQFL EAMESTGGRV RIAIAKLLSK QTS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
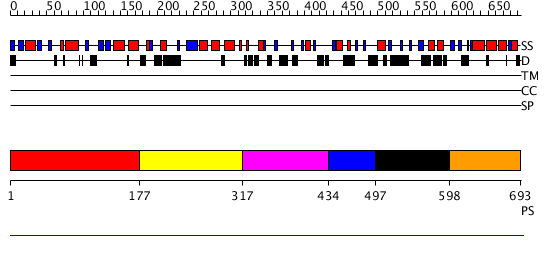
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..176] | 1.22 | c-Myb, DNA-binding domain | |
| 2 | View Details | [177..316] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [317..433] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [434..496] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [497..597] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [598..693] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)