













| General Information: |
|
| Name(s) found: |
rad4 /
SPAC23C4.18c
[Sanger Pombe]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 648 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA DNA replication preinitiation complex [ISS] spindle pole body [IDA nuclear replication fork [ISS spindle [IDA cytosol [IDA |
| Biological Process: |
DNA replication preinitiation complex assembly
[IMP DNA damage checkpoint [IMP DNA replication initiation [ISS DNA replication checkpoint [TAS S-M checkpoint [IMP |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSSKPLKGF VICCTSIDLK QRTEISTKAT KLGAAYRSDF TKDVTHLIAG DFDTPKYKFA 60
61 AKSRPDIKIM SSEWIPVLYE SWVQGEDLDD GLLVDKHFLP TLFKCRVCLT NIGQPERSRI 120
121 ENYVLKHGGT FCPDLTRDVT HLIAGTSSGR KYEYALKWKI NVVCVEWLWQ SIQRNAVLEP 180
181 QYFQLDMPAE KIGLGAYVRL DPNTTEAKSY SENQKISKNK EKSGQSLAAL AEEADLEPVI 240
241 MKRGKKRDRS ILWEELNNGK FEFSSRSEEN SVLLDDFTPE TVQPLEENEL DTELNIENEA 300
301 KLFKNLTFYL YEFPNTKVSR LHKCLSDNGG QISEFLSSTI DFVVIPHYFP VDELPIFSFP 360
361 TVNEWWIERC LYYKKIFGID EHALAKPFFR PSLVPYFNGL SIHLTGFKGE ELSHLKKALT 420
421 ILGAVVHEFL GVQRSILLVN TNEPFSMKTR FKIQHATEWN VRVVGVAWLW NIIQSGKFID 480
481 QVSPWAIDKK ENQEIKKFTN QNNMVFPTSD RDTRLQNSLA QQPIGHSTPH NSPSLLSVKK 540
541 RQNNHIRSNT LIQLNSNSKD STIFPRRSVT VPGDKIDTVW KSSVTKPETP TSPQEHVSYI 600
601 DPDAQREKHK LYAQLTSNVD AIPPANDLQN QENGLLLITE SHRKLRRR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
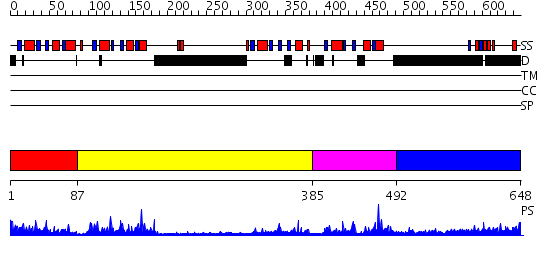
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | 39.522879 | No description for 2owoA was found. | |
| 2 | View Details | [87..384] | 12.69897 | Co-Crystal Structure of Lif1p-Lig4p | |
| 3 | View Details | [385..491] | 31.30103 | Breast cancer associated protein, BRCA1 | |
| 4 | View Details | [492..648] | 1.13899 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)