













| General Information: |
|
| Name(s) found: |
pyp2 /
SPAC19D5.01
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 711 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA mitochondrion [IDA cytosol [IDA |
| Biological Process: |
protein amino acid dephosphorylation
[IGI negative regulation of meiosis [IMP stress-activated MAPK cascade [IGI |
| Molecular Function: |
protein binding
[IPI protein tyrosine phosphatase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLHLLSKDEF NSTLKSFEEQ TESVSWIIDL RLHSKYAVSH IKNAINVSLP TALLRRPSFD 60
61 IGKVFACIKC NVKVSLDEIN AIFLYDSSMA GMNRIYDLVQ KFRRGGYSKK IYLLSNGFEA 120
121 FASSHPDAIV STEMVKESVP YKIDINENCK LDILHLSDPS AVSTPISPDY SFPLRVPINI 180
181 PPPLCTPSVV SDTFSEFASH AEYPGFSGLT PFSIHSPTAS SVRSCQSIYG SPLSPPNSAF 240
241 QAEMPYFPIS PAISCASSCP STPDEQKNFF IVGNAPQQTP ARPSLRSVPS YPSSNNQRRP 300
301 SASRVRSFSN YVKSSNVVNP SLSQASLEII PRKSMKRDSN AQNDGTSTMT SKLKPSVGLS 360
361 NTRDAPKPGG LRRANKPCFN KETKGSIFSK ENKGPFTCNP WGAKKVSPPP CEVLADLNTA 420
421 SIFYKFKRLE EMEMTRSLAF NDSKSDWCCL ASSRSTSISR KNRYTDIVPY DKTRVRLAVP 480
481 KGCSDYINAS HIDVGNKKYI ACQAPKPGTL LDFWEMVWHN SGTNGVIVML TNLYEAGSEK 540
541 CSQYWPDNKD HALCLEGGLR ISVQKYETFE DLKVNTHLFR LDKPNGPPKY IHHFWVHTWF 600
601 DKTHPDIESI TGIIRCIDKV PNDGPMFVHC SAGVGRTGTF IAVDQILQVP KNILPKTTNL 660
661 EDSKDFIFNC VNSLRSQRMK MVQNFEQFKF LYDVVDYLNS GVNQASKPLM T |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
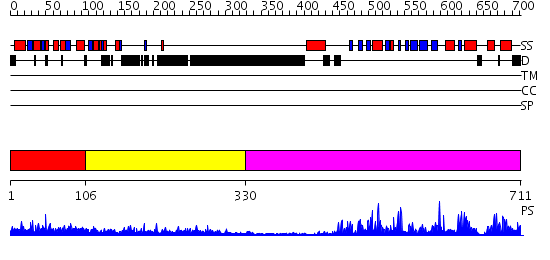
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..105] | 32.221849 | Erk2 binding domain of Mapk phosphatase mkp-3 | |
| 2 | View Details | [106..329] | 12.045757 | No description for 2v5yA was found. | |
| 3 | View Details | [330..711] | 87.0 | Crystal structure of human tyrosine phosphatase SHP-1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)