













| General Information: |
|
| Name(s) found: |
ade1 /
SPBC405.01
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 788 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
'de novo' IMP biosynthetic process
[IMP purine base biosynthetic process [IEA] |
| Molecular Function: |
ATP binding
[IEA]
phosphoribosylamine-glycine ligase activity [IMP manganese ion binding [IEA] phosphoribosylformylglycinamidine cyclo-ligase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEPIIALLIG NGGREHTIAW KLCESPLISK VYVAPGNGGT ASNGAESKME NVNIGVCDFE 60
61 QLVKFALDKD VNLVIPGPEL PLVEGIEGHF RRVGIPCFGP SALAARMEGS KVFSKDFMHR 120
121 NNIPTAVYKS FSNYDHAKSF LDTCTFDVVI KADGLAAGKG VIIPKTKKEA FEALESIMLN 180
181 EEFGSAGKNV VIEELLEGEE LSILTFSDGY TCRSLPPAQD HKRAFDGDKG PNTGGMGCYA 240
241 PTPVASPKLL ETVQSTIIQP TIDGMRHEGY PLVGILFTGL MLTPSGPRVL EYNVRFGDPE 300
301 TQAVLPLLES DLAEIILACV NHRLDAIDIV ISRKFSCAVV CVAGGYPGPY NKGDIITFDA 360
361 LKDKNTRIFH AGTSIRDGNV VTNGGRVLAV EATGDSVEAA VRLAYEGVKT VHFDKMFYRK 420
421 DIAHHALNPK RKTREILTYE NSGVSVDNGN EFVQRIKDLV KSTRRPGADA DIGGFGGIFD 480
481 LKQAGWNDPL LVSATDGVGS KLLIALSLNK HDTVGIDLVA MNVNDLVVQG AEPLIFLDYF 540
541 ATGSLDLKVS TSFVEGVVKG CKQAGCALVG GETSEMPGLY HDGHYDANGT SVGAVSRDDI 600
601 LPKPESFSKG DILLGLASDG VHSNGYSLVR KIVEYSDLEY TSVCPWDKNV RLGDSLLIPT 660
661 RIYVKPLLHV IRKNIVKGMA HITGGGLVEN VPRMLPSHLN AIIDVDTWEV PEVFKWLKDA 720
721 GNVPISDMAR TFNMGIGMVV AVASEDAEET MKELTSVGET VYRIGQLVDK ESSSERCHLV 780
781 NLNKWETF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
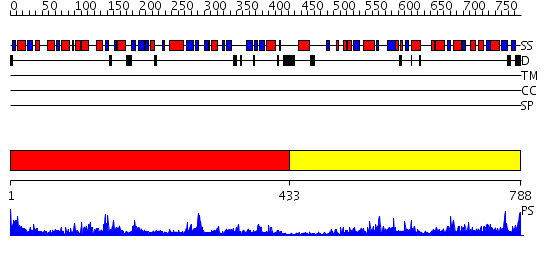
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..432] | 93.522879 | No description for 2qk4A was found. | |
| 2 | View Details | [433..788] | 83.69897 | Aminoimidazole ribonucleotide synthetase (PurM) N-terminal domain; Aminoimidazole ribonucleotide synthetase (PurM) C-terminal domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)