













| General Information: |
|
| Name(s) found: |
nif1 /
SPBC23G7.04c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 681 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cell tip [IDA cytosol [IDA |
| Biological Process: |
negative regulation of G2/M transition of mitotic cell cycle
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METMQSRTYA GLTKLNTTTA LLNKKDGNDD DKAEHSKRSG YHGLSPLDAL ALKHRDLNRK 60
61 LNLRAMMSKS EDNLQILKET TSGSSSDLLN IESPASPAEA SSPFTVRTPT VHDPEHYFVA 120
121 QKLSSVFGTP DLEDETDFFD YFSAAPDVHP NDIFDSYNSN NIAESFDDDN YYNSLLPPNA 180
181 PYYHEIEPPR TASNTSPTPN SIKSAHPAEP PKRPAFTRSA TSPDKILPTR IKSKDTVSSG 240
241 DSTPLSGSSS SKGMLMSMST SENHSLSSNP ELSNSNLLAK NESPADVSNN ESGNESSKEP 300
301 DKEHSTPIHP TTPVSRCARP SSRQQTISIL QAQSPFLKKS DKERANLNKT MVSINKSINI 360
361 HQSIHEISCP HHSSSDNCLF ILISLMDRLH SPVLKQLDVS LQSLTMTAIR YIDLNYVDVQ 420
421 YTNLRGGAYG NGNNSESSDN AQLKKEEHLN LAIHFHLLND HDKCFWHTGM ASSYEDYTAT 480
481 FIYGLYLRHG LACSPKTHVS FLFLLKTATQ LLNKLVECLH SSDLGLNDTT PNEKLSTEYN 540
541 QQRLLLALIL YELGVCFMHG WGITRDRYLA LHLIKLSGAW GDADAQFEAG LQMSLGAVSD 600
601 KDSHMAAYYY RLAGFQGISP PSKCKWVYKS KYSLAANHKV PAASEVAYVS AIVENLESHS 660
661 LKFSTKPKAK LRSLITSVRY L |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
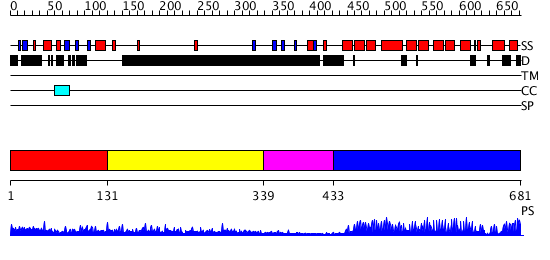
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..130] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [131..338] | 1.091994 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [339..432] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [433..681] | 63.045757 | Helicobacter cysteine rich protein C (HcpC) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.58 |
Source: Reynolds et al. (2008)