













| General Information: |
|
| Name(s) found: |
ned1 /
SPAC1952.13
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 656 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[ISS cytosol [IDA |
| Biological Process: |
ascospore formation
[IMP [NOT]nucleocytoplasmic transport [IMP nuclear envelope organization [ISS chromosome organization [IMP phospholipid biosynthetic process [ISS cellular response to stress [IEP endoplasmic reticulum organization [IMP dephosphorylation [IC nucleus organization [IMP |
| Molecular Function: |
magnesium ion binding
[ISS phosphatidate phosphatase activity [ISS protein binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQYVGRAFDS VTKTWNAINP STLSGAIDVI VVEQEDKTLA CSPFHVRFGK FSLLLPSDKK 60
61 VEFSVNGQLT GFNMKLGDGG EAFFVFATEN AVPRELQTSP IVSPTTSPKQ TPSINVTEPQ 120
121 DLELDKVSQD HEKDQSNTYL MEDGYEFPLT RDLIRRSKSD ADQTPPTGFK HLRHSSCLEM 180
181 AGSDRTPSMP ATTLADLRLL QKAKELGKRL SGKELPTRVG DNGDVMLDMT GYKSSAANIN 240
241 IAELARETFK DEFPMIEKLL REDEEGNLWF HASEDAKKFA EVYGHSPPAS PSRTPASPKS 300
301 DSALMDEDSD LSRRHSLSEQ SLSPVSESYP QYAKTLRLTS DQLRSLNLKP GKNELSFGVN 360
361 GGKAICTANL FFWKHNDPVV ISDIDGTITK SDALGHMFTL IGKDWTHAGV AKLYTDITNN 420
421 GYKIMYLTSR SVGQADSTRH YLRNIEQNGY SLPDGPVILS PDRTMAALHR EVILRKPEVF 480
481 KMACLRDLCN IFALPVPRTP FYAGFGNRIT DAISYNHVRV PPTRIFTINS AGEVHIELLQ 540
541 RSGHRSSYVY MNELVDHFFP PIEVSTRDEV SSFTDVNFWR SPLLELSDEE EDDTNKSTSK 600
601 SPKTPKNTKF GYQEFEGIDE EDAQDYSPSP LIKSFNELMF EGEEDEEGEE DVENAV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
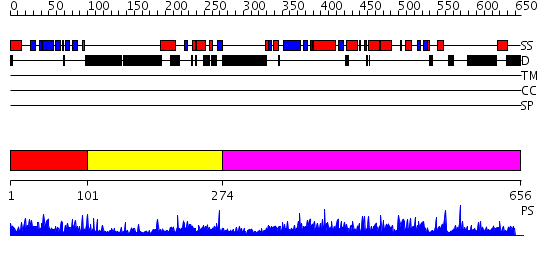
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..100] | 68.408935 | No description for PF04571.5 was found. Confident ab initio structure predictions are available. | |
| 2 | View Details | [101..273] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [274..656] | 1.24 | No description for 2hlkA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)