













| General Information: |
|
| Name(s) found: |
hal4 /
SPAC29A4.16
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 636 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
cellular homeostasis
[NAS]
regulation of membrane potential [IMP response to drug [IMP response to cation stress [IGI protein amino acid phosphorylation [ISS cellular cation homeostasis [IGI |
| Molecular Function: |
ATP binding
[ISS protein binding [IPI protein serine/threonine kinase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGEKDKLHEI SSKFASLGLG SLKSTPKARE TTEPPPPSSQ QPPSTPNGKE AASPSALKQN 60
61 VRPSLNSVQQ TPASIDAVAS SSNVSLQSQQ PLSKPVVSSK PNQTTAMPPP SNNPSRHVSS 120
121 TSNKPAAVSP NPAAHHAELP SGSVPPSASV SRANSTATTT PHKAGVVSNP AAANVHVLSV 180
181 AASPNPSTPS NGPAPVSTTA TPSRNPVTRL QRIFSQNSVS RQNSRTGRGA AVANTEETNS 240
241 TGGSETGGAA NSSSTSNPSS AKWSRFTVYD DASHTHQLRP ARRQEKLGKM LKDFLAGNSK 300
301 KREEERIAKE AADAQHQLSL VQSWINGYGQ EKLADKKDPA KVSASFVEKY GRCQEVIGRG 360
361 AFGVVRIAHK VDPQNSGSET LYAVKEFRRK PAESQKKYTK RLTSEFCISS SLRHPNVIHT 420
421 LDLIQDGKGD YCEVMELCSG GDLYTLIMAA GRLEPMEADC FFKQLMRGVD YLHDMGVAHR 480
481 DLKPENLLLT VSGSLKITDF GNGECFRMAW EKEAHMTCGL CGSAPYIAPE EYTESEFDPR 540
541 AVDVWACGVI YMAMRTGRHL WRVAKKSEDE YYSRYLMDRK NESGYEPIEM LERSRCRNTL 600
601 YNILHPNPTY RLTAKQIMKS EWVRSITLCE AGNAGL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
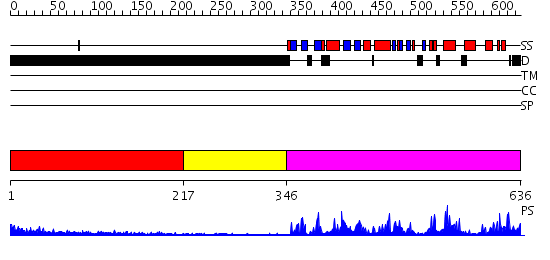
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..216] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [217..345] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [346..636] | 75.221849 | No description for 2fh9A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)