













| General Information: |
|
| Name(s) found: |
hob1 /
SPBC21D10.12
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 466 amino acids |
Gene Ontology: |
|
| Cellular Component: |
medial cortex
[TAS cell cortex of cell tip [TAS |
| Biological Process: |
chromatin silencing at centromere
[IMP establishment or maintenance of cell polarity [IGI cellular response to stress [IMP telomere maintenance [IGI response to DNA damage stimulus [IGI chromatin silencing at telomere [IMP regulation of mitotic cell cycle [IMP |
| Molecular Function: |
protein binding
[IPI GTPase binding [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSWKGFTKAL ARTPQTLRSK FNVGEITKDP IYEDAGRRFK SLETEAKKLA EDAKKYTDAI 60
61 NGLLNHQIGF ADACIEIYKP ISGRASDPES YEQEGNAEGI EAAEAYKEIV YDLQKNLASE 120
121 MDVINTRIVN PTGELLKIVK DVDKLLLKRD HKQLDYDRHR SSFKKLQEKK DKSLKDEKKL 180
181 YEAETAFEQS SQEYEYYNEM LKEELPKLFA LAQSFIAPLF QGFYYMQLNV YYVLYEKMSH 240
241 CEIQYFDFNT DILESYERRR GDVKDRAEAL TITKFKTAKP TYKRPGMGPG GKDATASSSS 300
301 SFSSKREEAA AEPSSSTATD IPPPYSTPSV AGASDYSTPS AGYQTVQTTT TTTEAAAAQY 360
361 PQAAFPPPPV MPQPAAAAVT TPVAAPVAAA AAAVPVPPPA PAPAAAPAAE HVVALYDYAA 420
421 QAAGDLSFHA GDRIEVVSRT DNQNEWWIGR LNGAQGQFPG NYVQLE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
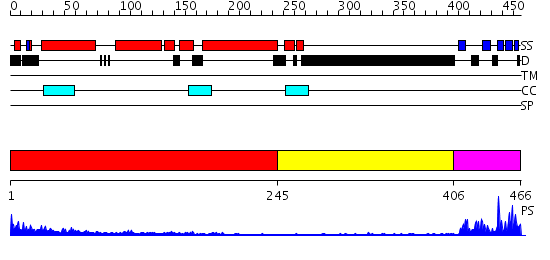
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..244] | 23.522879 | No description for 2ficA was found. | |
| 2 | View Details | [245..405] | 1.26 | No description for 2j63A was found. | |
| 3 | View Details | [406..466] | 19.39794 | Crystal structure of the SH3 domain from a S. cerevisiae hypothetical 40.4 kDa protein at 1.39 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)