













| General Information: |
|
| Name(s) found: |
hmt2 /
SPBC2G5.06c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 459 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
response to cadmium ion
[IMP regulation of sulfur metabolic process [IMP cellular cadmium ion homeostasis [IMP |
| Molecular Function: |
cadmium ion binding
[IEA]
oxidoreductase activity, acting on sulfur group of donors, quinone or similar compound as acceptor [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLTLNSTIKS VTGSFQSASM LARFASTHHK VLVVGGGSAG ISVAHQIYNK FSKYRFANDQ 60
61 GKDTSLKPGE IGIVDGAKYH YYQPGWTLTG AGLSSVAKTR RELASLVPAD KFKLHPEFVK 120
121 SLHPRENKIV TQSGQEISYD YLVMAAGIYT DFGRIKGLTE ALDDPNTPVV TIYSEKYADA 180
181 VYPWIEKTKS GNAIFTQPSG VLKCAGAPQK IMWMAEDYWR RHKVRSNIDV SFYTGMPTLF 240
241 SVKRYSDALL RQNEQLHRNV KINYKDELVE VKGSERKAVF KNLNDGSLFE RPFDLLHAVP 300
301 SMRSHEFIAK SDLADKSGFV AVDQSTTQST KFPNVFAIGD CSGLPTSKTY AAITAQAPVM 360
361 VHNLWSFVNG KNLTASYNGY TSCPLLTGYG KLILAEFLYK QEPKESFGRF SRFLDQTVPR 420
421 RMFYHLKKDF FPFVYWNFAV RNGKWYGSRG LIPPHFPVN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
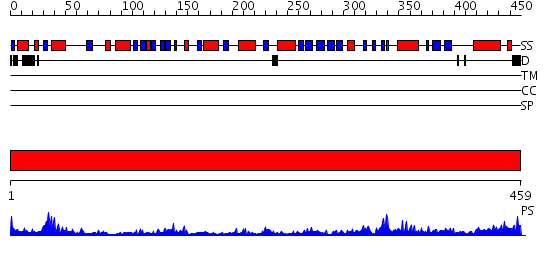
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..459] | 51.522879 | Dihydrolipoamide dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.44 |
Source: Reynolds et al. (2008)