













| General Information: |
|
| Name(s) found: |
gap1 /
SPBC646.12c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 766 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
regulation of small GTPase mediated signal transduction
[IEA]
regulation of catalytic activity [IC Ras protein signal transduction [IC regulation of conjugation with cellular fusion by signal transduction [IMP |
| Molecular Function: |
Ras GTPase activator activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTKRHSGTLS SSVLPQTNRL SLLRNRESTS VLYTIDLDME SDVEDAFFHL DRELHDLKQQ 60
61 ISSQSKQNFV LERDVRYLDS KIALLIQNRM AQEEQHEFAK RLNDNYNAVK GSFPDDRKLQ 120
121 LYGALFFLLQ SEPAYIASLV RRVKLFNMDA LLQIVMFNIY GNQYESREEH LLLSLFQMVL 180
181 TTEFEATSDV LSLLRANTPV SRMLTTYTRR GPGQAYLRSI LYQCINDVAI HPDLQLDIHP 240
241 LSVYRYLVNT GQLSPSEDDN LLTNEEVSEF PAVKNAIQER SAQLLLLTKR FLDAVLNSID 300
301 EIPYGIRWVC KLIRNLTNRL FPSISDSTIC SLIGGFFFLR FVNPAIISPQ TSMLLDSCPS 360
361 DNVRKTLATI AKIIQSVANG TSSTKTHLDV SFQPMLKEYE EKVHNLLRKL GNVGDFFEAL 420
421 ELDQYIALSK KSLALEMTVN EIYLTHEIIL ENLDNLYDPD SHVHLILQEL GEPCKSVPQE 480
481 DNCLVTLPLY NRWDSSIPDL KQNLKVTRED ILYVDAKTLF IQLLRLLPSG HPATRVPLDL 540
541 PLIADSVSSL KSMSLMKKGI RAIELLDELS TLRLVDKENR YEPLTSEVEK EFIDLDALYE 600
601 RIRAERDALQ DVHRAICDHN EYLQTQLQIY GSYLNNARSQ IKPSHSDSKG FSRGVGVVGI 660
661 KPKNIKSSNT VKLSSQQLKK ESVLLNCTIP EFNVSNTYFT FSSPSTDNFV IAVYQRGHSK 720
721 VLVEVCICLD DVLQRRYASN PVVDLGFLTF EANKLYHLFE QLFLRK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
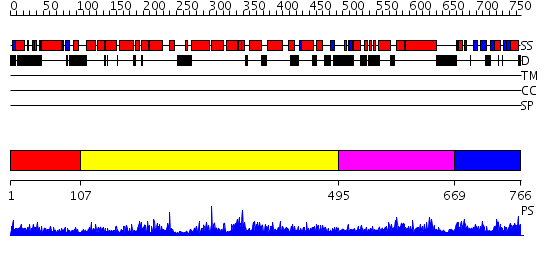
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 10.0 | No description for 2uzpA was found. | |
| 2 | View Details | [107..494] | 67.0 | GAP related domain of neurofibromin | |
| 3 | View Details | [495..668] | 35.327902 | No description for PF03836.6 was found. No confident structure predictions are available. | |
| 4 | View Details | [669..766] | 33.0 | Solution structure of the carboxyl-terminal RGC domain in human IQGAP1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.44 |
Source: Reynolds et al. (2008)