













| General Information: |
|
| Name(s) found: |
SPBPB2B2.12c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 713 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
carbohydrate metabolic process
[IEA]
cellular response to stress [IEP galactose metabolic process [IEA] |
| Molecular Function: |
coenzyme binding
[IEA]
UDP-glucose 4-epimerase activity [IEA] aldose 1-epimerase activity [IEA] carbohydrate binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVQDEYILV TGGAGYIGSH TVIELINHGY KVIIVDNLCN SCYDAVARVE FIVRKSIKFF 60
61 KLDLRDKEGL AQIFDTFKIK GVIHFAALKA VGESMKLPLE YYDNNICGTI TLLNVMREHR 120
121 VKTVVFSSSA TVYGDATRFD NMIPIPESCP NDPTNPYGKT KYAIENIIKD LHTSDNTWRG 180
181 AILRYFNPIG AHPSGLLGED PLGIPNNLLP FLAQVAIGRR EKLLVFGDDY DSHDGTPIRD 240
241 YIHVVDLAKG HIAALNYLNK INNSEGMYRE WNLGTGKGSS VFDIYHAFCK EVGKDLPYEV 300
301 VGRRTGDVLN LTASPNRANS ELKWKAELSI TDACRDLWKW TIENPFGFQI DNYKWKLFNT 360
361 LGIMDYKNRL HTICFQDLEV SIANYGALVQ AVRYKGRNLV NGFNDFSRYK LKENPFFGAT 420
421 IGRFANRIAN GQFEVDGHLY TLCKNENNKT TLHGGNNGFD KQFFLGPIAR QYEDYNTLEF 480
481 ILVDKDGNNG FPSDLETLVK YTIKNNSLEI EYKSVIPEYS KLNVTAVNLT NHSYWNLASP 540
541 NKTIDGTIIK STTNVYLKVN SETSLPTGDI VEWQNDITKP TKLDPNISFD NCFIVDREAS 600
601 KFCLDTRKYS LKNIVEVIHP SVPVKLVVST TEPAFQLYTG DGNDICEFQS RSGFCVETGR 660
661 FINALNNEKW SKQVILRKGE VYGARSKFSL YAQDLEENKH FLDSASYNSG EYY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
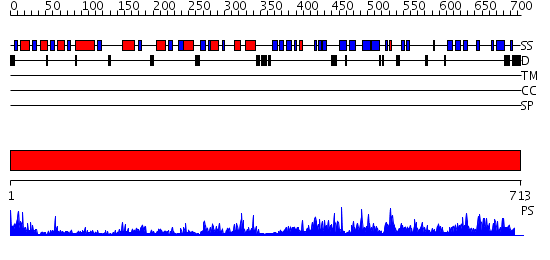
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..713] | 171.0 | Crystal structure of the gal10 fusion protein galactose mutarotase/UDP-galactose 4-epimerase from Saccharomyces cerevisiae complexed with NAD, UDP-glucose, and galactose |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)