













| General Information: |
|
| Name(s) found: |
eso1 /
SPBC16A3.11
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 872 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IC] |
| Biological Process: |
DNA repair
[ISS regulation of maintenance of mitotic sister chromatid cohesion [TAS DNA-dependent DNA replication [IMP establishment of mitotic sister chromatid cohesion [IMP protein amino acid acetylation [IC fear response [IC] translesion synthesis [ISS |
| Molecular Function: |
DNA-directed DNA polymerase activity
[ISS zinc ion binding [IEA] protein binding [IPI acyltransferase activity [ISS damaged DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MELGKSKFSW KDLQYCDKAG TQNSPLRVVA HIDQDAFYAQ VESVRLGLDH SVPLAVQQWQ 60
61 GLIAVNYAAR AANISRHETV TEAKKKCPEL CTAHVKTWKA GESEAKYHEN PNPNYYKTCL 120
121 DPYRHESVKI LNIIKKHAPV VKKASIDECF IELTSDVKRI VLEEYPYLKI PSEDSNVALP 180
181 QAPVLLWPAE FGMVIEEEVV DRTKEDYERD WDDVFLFYAA KIVKEIRDDI YLQLKYTCSA 240
241 GVSFNPMLSK LVSSRNKPNK QTILTKNAIQ DYLVSLKITD IRMLGGKFGE EIINLLGTDS 300
301 IKDVWNMSMD FLIDKLGQTN GPLVWNLCHG IDNTEITTQV QIKSMLSAKN FSQQKVKSEE 360
361 DAINWFQVFA SDLRSRFLEL EGMRRPKTIC LTVVSRFLRK SRSSQIPMNV DISTQFIVEA 420
421 TSKLLRQLQQ EFDVYPISNL SISFQNIIEV DRNSRGIEGF LKKSNDEIYM STSVSPSIEG 480
481 RAKLLNENMR ENNSFELSSE KDIKSPKRLK RGKGKGIFDM LQQTAVSKPT ENSADETYTC 540
541 EECEQKITLS ERNEHEDYHI ALSISRKERY NNLVPPSHDK PKQVKPKTYG RKTGSKHYAP 600
601 LSDETNNKRA FLDAFLGNGG NLTPNWKKQT PKAISNSSDN MTQLHLDLAN STVTCSECSM 660
661 EYNSTSEEDI LLHSRFHSRV LGGVTVSFQC SPIYRVNYGL SSDCIYSINS ESSLIDQRKA 720
721 EEALSFVNNE LSSEPIETIG VDKYTTFLFI SDKKCVGLLL AERISSAYIV DELELNNNNS 780
781 TSSAVYIKNE NLRKGFVLGI SRIWVSASRR KQGIASLLLD NALKKFIYGY VISPAEVAFS 840
841 QPSESGKQFI ISWHRSRNNG SSKSLRYAVY ES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
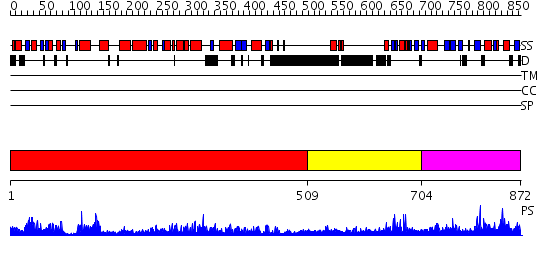
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..508] | 71.0 | DNA polymerase eta | |
| 2 | View Details | [509..703] | 2.42 | No description for 2i13A was found. | |
| 3 | View Details | [704..872] | 3.060963 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)