













| General Information: |
|
| Name(s) found: |
mid1 /
SPCC4B3.15
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 920 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA nucleus [IDA medial cortex [IDA medial cortical node [IDA |
| Biological Process: |
protein homooligomerization
[IDA cytokinesis [IMP cellular protein localization [IGI cytokinesis, site selection [IMP |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKEQEFSYRE AKDVSLDSKG LENSFLSSPN REKTPLFFEG NSNETSGYDQ TKNFTHGDGD 60
61 MSLGNLSELN VATDLLESLD LRSMYMHGYG HLDSSFSSQH SPDNRKRMSS TSVFKRINSE 120
121 EEGRIPSLTY SAGTMNSTSS STASLKGADI VADYETFNPD QNLAELSFDR SKSSRKRAVE 180
181 VAEFSRAKTM SPLEYTVQHP YQSHNELSTN PARARAGSVP NLARIPSDVK PVPPAHLSAS 240
241 STVGPRILPS LPKDTTEDNP ALERVETTAS LDMDYKPLEP LAPIQEAPVE DTSEPFSSVP 300
301 EATLDDSDIS TESLRKKVLA KMEAKRISSG SSYASTLRKV YDFSELSLPT NGKDYDELYL 360
361 QSSRNSEPEI STIINDSLQQ ENMDEDISAT SIPKSQAAYG HGSVTYHEVP RYNLTSASVG 420
421 YSISSQRGRI KSSSTIDNLS AILSSEDLRH PSMQPVPGTK RTYSNYCENE PNKSSQSLVS 480
481 SESHNVEGWN YSETGTVGFY DPSAEISASI DELRQSTPVA RDSELLSRAH SFDLNRLDLP 540
541 SQDKSTSYEV PNGTENQSPR PVTSLGFVNE TFFEEKPKAP LPLGRFYIHL NSILNISISE 600
601 VHSPIKIIVN TPTQNMQLPW QAVNGNNRLD HDFAFHVDDN FKVSFMFLDI PIEDKSNGSK 660
661 GVSATKDVSN GKPAETKSKA RKFFDKLFNR RKKRKLNKAA AVENSKAKKS VVIKKVSGTA 720
721 TLNLGNVKDS CFGKAFNVEI PIISRGFLEA IPVKINSIGK RTLGNLTLTC LYIPELSVPE 780
781 QELPFTLEQA TMDLRHVRSN YLYNEGYLYR LEDSSIRRRF VVLRSKQLNF YAEKGGQYLD 840
841 TFQLSKTVVS IPMVNFSEAV SNLGLVAGIL ATSVDRRHVQ LFADSKKVCQ KWLQVMNSRS 900
901 FALDRGTEKL WLQEYVNFMA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
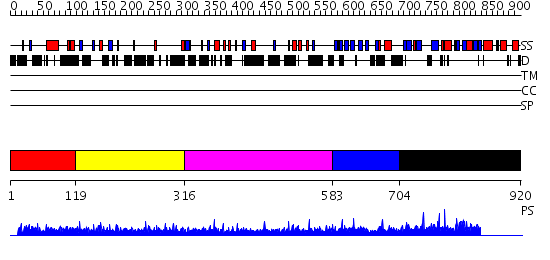
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [119..315] | 4.215997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [316..582] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [583..703] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [704..920] | 1.28 | Crystal Structure of the PDK1 Pleckstrin Homology (PH) domain bound to Inositol (1,3,4,5)-tetrakisphosphate |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)