













| General Information: |
|
| Name(s) found: |
prp22 /
SPAC10F6.02c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1168 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA spliceosomal complex [ISS |
| Biological Process: |
RNA splicing, via transesterification reactions
[ISS spliceosome disassembly [ISS |
| Molecular Function: |
ATP binding
[IEA]
RNA binding [IEA] ATP-dependent RNA helicase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDDLKELEYL SLVSKVASEI RNHTGIDDNT LAEFIINLHD QSKNYDEFKN NVLSCGGEFT 60
61 DSFLQNISRL IKEIKPKDDI PTDNVNNGSN SVNGASHDLD SKDVDKQHQR KMFPGLSIPN 120
121 STNNLRDRPA LMDNAMDELE ELSTLAKTRR NDRDSRRDER HYLNGIRERR ERSISPSFSH 180
181 HSRTSISGQS HSSRSSRGPL LNAPTLYGIY SGVVSGIKDF GAFVTLDGFR KRTDGLVHIS 240
241 NIQLNGRLDH PSEAVSYGQP VFVKVIRIDE SAKRISLSMK EVNQVTGEDL NPDQVSRSTK 300
301 KGSGANAIPL SAQNSEIGHV NPLETFTSNG RKRLTSPEIW ELQQLAASGA ISATDIPELN 360
361 DGFNTNNAAE INPEDDEDVE IELREEEPGF LAGQTKVSLK LSPIKVVKAP DGSLSRAAMQ 420
421 GQILANDRRE IRQKEAKLKS EQEMEKQDLS LSWQDTMSNP QDRKFAQDVR DSAARQLTSE 480
481 TPSWRQATRN ANISYGKRTT LSMKEQREGL PVFKLRKQFL EAVSKNQILV LLGETGSGKT 540
541 TQITQYLAEE GYTSDSKMIG CTQPRRVAAM SVAKRVAEEV GCRVGEEVGY TIRFEDKTSR 600
601 MTQIKYMTDG MLQRECLVDP LLSKYSVIIL DEAHERTVAT DVLFGLLKGT VLKRPDLKLI 660
661 VTSATLDAER FSSYFYKCPI FTIPGRSYPV EIMYTKQPEA DYLDAALMTV MQIHLSEGPG 720
721 DILVFLTGQE EIDTSCEILY ERSKMLGDSI PELVILPVYS ALPSEIQSRI FEPAPPGGRK 780
781 VVIATNIAET SLTIDGIYYV VDPGFVKQSC FDPKLGMDSL IVTPISQAQA RQRSGRAGRT 840
841 GPGKCYRLYT ESAYRNEMLP SPIPEIQRQN LSHTILMLKA MGINDLLNFD FMDPPPAQTM 900
901 IAALQNLYAL SALDDEGLLT PLGRKMADFP MEPQLSKVLI TSVELGCSEE MLSIIAMLSV 960
961 PNIWSRPREK QQEADRQRAQ FANPESDHLT LLNVYTTWKM NRCSDNWCYE HYIQARGMRR1020
1021 AEDVRKQLIR LMDRYRHPVV SCGRKRELIL RALCSGYFTN VAKRDSHEGC YKTIVENAPV1080
1081 YMHPSGVLFG KAAEWVIYHE LIQTSKEYMH TVSTVNPKWL VEVAPTFFKF ANANQVSKTK1140
1141 KNLKVLPLYN RFEKPDEWRI SKQRKGGR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
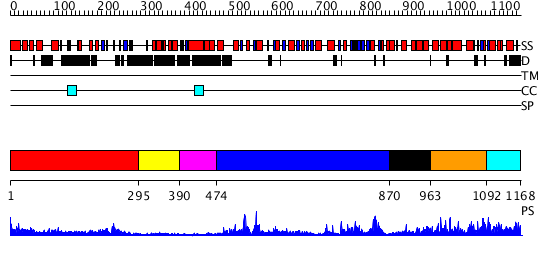
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..294] | 15.09691 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 2 | View Details | [295..389] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [390..473] | 4.522879 | Crystal structure of the UvsW helicase from Bacteriophage T4 | |
| 4 | View Details | [474..869] | 51.522879 | Initiation factor 4a | |
| 5 | View Details | [870..962] | 12.39794 | Nucleotide excision repair enzyme UvrB | |
| 6 | View Details | [963..1091] | N/A | Confident ab initio structure predictions are available. | |
| 7 | View Details | [1092..1168] | 1.238997 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)