













| General Information: |
|
| Name(s) found: |
ctf1 /
SPBC3B9.11c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 363 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mRNA cleavage and polyadenylation specificity factor complex [IDA cytosol [IDA |
| Biological Process: |
mRNA polyadenylation
[IC |
| Molecular Function: |
RNA binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSMTAGNVVF VGNIPYDVSE QQMTEIFNQV GPVKTFKLVL DPETGSGKGY GFCEFFDSET 60
61 TAMAVRKLNN SELGPRKIRV EFPSNDPRRN QSYEYTERTD RYMEQQNAHE SSYNSRFIPP 120
121 VLHSTSSLPA SQGGGMPSPA IYSSSMATNL NKNINSTSVP AYNFHNSMTS DFDSASQPHT 180
181 DAYNARTFQY NKSSQNKGDY TSGTSISNPT SIPLAPSVVQ VLSTFSAQEL LNMLSKLQTV 240
241 VHIAPEEARR LLIANPALPY AAFQAMLLMN LVDANVLQQV VVAVKNKNMH QPASATSSPP 300
301 SVPQKIPSSN HKSQQANGSD QGNEGKRMAL IQQLLALTPE QINALPPAQR DQILSIRRQH 360
361 FRQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
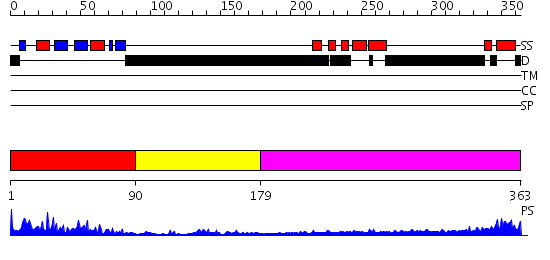
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | 22.39794 | NMR Structure of the N-terminal RRM domain of Cleavage stimulation factor 64 KDa subunit | |
| 2 | View Details | [90..178] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [179..363] | 5.39794 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)