













| General Information: |
|
| Name(s) found: |
cap1 /
SPCC306.09c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 551 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA actin cortical patch [ISS |
| Biological Process: |
ascospore formation
[IMP cytoskeleton organization [ISS regulation of adenylate cyclase activity [IGI conjugation with cellular fusion [IMP cellular response to starvation [IMP |
| Molecular Function: |
actin binding
[NAS adenylate cyclase binding [IPI cytoskeletal protein binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDMINIRET GYNFTTILKR LEAATSRLED LVESGHKPLP NMHRPSRDSN SQTHNISFNI 60
61 GTPTAPTVST GSPAVASLHD QVAAAISPRN RSLTSTSAVE AVPASISAYD EFCSKYLSKY 120
121 MELSKKIGGL IAEQSEHVEK AFNLLRQVLS VALKAQKPDM DSPELLEFLK PIQSELLTIT 180
181 NIRDEHRTAP EFNQLSTVMS GISILGWVTV EPTPLSFMSE MKDSSQFYAN RVMKEFKGKD 240
241 DLQIEWVRSY LTLLTELITY VKTHFKTGLT WSTKQDAVPL KTALANLSAS KTQAPSSGDS 300
301 ANGGLPPPPP PPPPSNDFWK DSNEPAPADN KGDMGAVFAE INKGEGITSG LRKVDKSEMT 360
361 HKNPNLRKTG PTPGPKPKIK SSAPSKPAET APVKPPRIEL ENTKWFVENQ VDNHSIVLDS 420
421 VELNHSVQIF GCSNCTIIIK GKLNTVSMSN CKRTSVVVDT LVAAFDIAKC SNFGCQVMNH 480
481 VPMIVIDQCD GGSIYLSKSS LSSEVVTSKS TSLNINVPNE EGDYAERAVP EQIKHKVNEK 540
541 GELVSEIVRH E |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
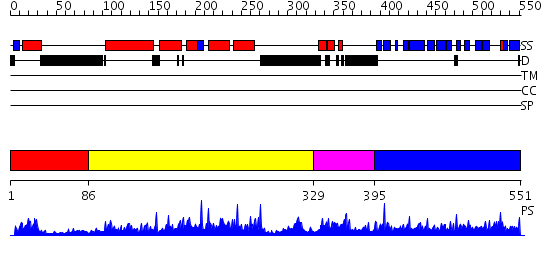
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | 1.039994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [86..328] | 67.69897 | The crystal structure of the N-terminal domain of CAP indicates variable oligomerisation | |
| 3 | View Details | [329..394] | 1.056988 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [395..551] | 76.221849 | C-terminal domain of adenylylcyclase associated protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)