













| General Information: |
|
| Name(s) found: |
Fim-PC /
FBpp0074216
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 616 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
female meiosis chromosome segregation
[IMP]
|
| Molecular Function: |
actin binding
[IEA][ISS][NAS]
calcium ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLDANKDGFI DLHELKDALN QVGFKLAGYQ VREMIDEYKG KQLTAFQGKL NLEEFEALCL 60
61 DLKSKDVAST FKTVVSKKEN LETLGGMSSI SSEGTTHSVR LEEQLAFSDW INSNLGHDKD 120
121 LQHLLPIDSE GKRLYLSIKD GILLCKIINH SCPDTIDERA INKKNLTVYR EFENLTLALV 180
181 SSQAIGCNIV NIDAHDLAKG KPHLVLGLLW QIIRIGLFSH ITLDSCPGLA GLLFDNERLE 240
241 DLMKMSPEAI LLRWVNHHLE RAGISRRCTN FQSDIVDSEI YSHLLKQIAG NDADVNLDAL 300
301 RESDLQSRAE IMLQQAAKLN CRSFLTPQDV VNGVYKLNLA FVANLFNNHP GLDKPEQIEG 360
361 LESIEETREE KTYRNWMNSM GVAPHVNWLY SDLADGLVIF QLFDVIKPGI VNWSRVHKRF 420
421 SPLRKFMEKL ENCNYAVDLG KQLKFSLVGI AGQDLNDGNA TLTLALIWQL MRAYTLSILS 480
481 RLANTGNPII EKEIVQWVNN RLSEAGKQSQ LRNFNDPAIA DGKIVIDLID AIKEGSINYE 540
541 LVRTSGTQED NLANAKYAIS MARKIGARVY ALPEDITEVK PKMVMTVFAC MMALDYVPNM 600
601 DSVDQNNHNS SANNSN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
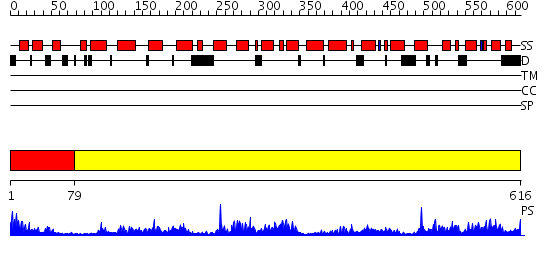
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | 16.522879 | THE LINKER OF DES-GLU84 CALMODULIN IS BENT AS SEEN IN THE CRYSTAL STRUCTURE | |
| 2 | View Details | [79..616] | 114.0 | CRYSTAL STRUCTURE OF THE ACTIN-CROSSLINKING CORE OF SCHIZOSACCHAROMYCES POMBE FIMBRIN |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)