













| General Information: |
|
| Name(s) found: |
baz-PA /
FBpp0074162
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1464 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apicolateral plasma membrane
[TAS]
adherens junction [IEP] subapical complex [TAS] apical part of cell [NAS] spot adherens junction [IDA] cell-cell adherens junction [NAS] cytoplasm [NAS] cell cortex [TAS] apical junction complex [IEP] apical cortex [NAS][TAS] apical plasma membrane [NAS][TAS] |
| Biological Process: |
establishment of apical/basal cell polarity
[IMP]
germarium-derived oocyte fate determination [IMP] protein localization [TAS] germ-band extension [IMP][TAS] border follicle cell delamination [IMP] cytokinesis [IMP] border follicle cell migration [IMP] ovarian follicle cell-cell adhesion [IMP] asymmetric protein localization [TAS] establishment or maintenance of neuroblast polarity [NAS] oocyte axis specification [TAS] basal protein localization [TAS] establishment or maintenance of epithelial cell apical/basal polarity [IMP][NAS][TAS] apical protein localization [NAS] establishment or maintenance of polarity of embryonic epithelium [NAS] establishment of mitotic spindle localization [TAS] cell-cell junction assembly [NAS] establishment or maintenance of cell polarity [NAS] asymmetric cell division [TAS] synapse assembly [IMP] morphogenesis of an epithelium [NAS][TAS] asymmetric protein localization involved in cell fate determination [IMP] zonula adherens assembly [IMP][NAS][TAS] morphogenesis of a polarized epithelium [TAS] asymmetric neuroblast division [IGI][IMP] |
| Molecular Function: |
protein kinase C binding
[ISS]
protein binding [IPI][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKVTVCFGDV RILVPCGSGE LLVRDLVKEA TRRYIKAAGK PDSWVTVTHL QTQSGILDPD 60
61 DCVRDVADDR EQILAHFDDP GPDPGVPQGG GDGASGSSSV GTGSPDIFRD PTNTEAPTCP 120
121 RDLSTPHIEV TSTTSGPMAG LGVGLMVRRS SDPNLLASLK AEGSNKRWSA AAPHYAGGDS 180
181 PERLFLDKAG GQLSPQWEED DDPSHQLKEQ LLHQQQPHAA NGGSSSGNHQ PFARSGRLSM 240
241 QFLGDGNGYK WMEAAEKLQN QPPAQQTYQQ GSHHAGHGQN GAYSSKSLPR ESKRKEPLGQ 300
301 AYESIREKDG EMLLIINEYG SPLGLTALPD KEHGGGLLVQ HVEPGSRAER GRLRRDDRIL 360
361 EINGIKLIGL TESQVQEQLR RALESSELRV RVLRGDRNRR QQRDSKVAEM VEVATVSPTR 420
421 KPHAAPVGTS LQVANTRKLG RKIEIMLKKG PNGLGFSVTT RDNPAGGHCP IYIKNILPRG 480
481 AAIEDGRLKP GDRLLEVDGT PMTGKTQTDV VAILRGMPAG ATVRIVVSRQ QELAEQADQP 540
541 AEKSAGVAVA PSVAPPAVPA AAAPAPPIPV QKSSSARSLF THQQQSQLNE SQHFIDAGSE 600
601 SAASNDSLPP SSNSWHSREE LTLHIPVHDT EKAGLGVSVK GKTCSNLNAS GSSASSGSNG 660
661 LMKHDGDLGI FVKNVIHGGA ASRDGRLRMN DQLLSVNGVS LRGQNNAEAM ETLRRAMVNT 720
721 PGKHPGTITL LVGRKILRSA SSSDILDHSN SHSHSHSNSS GGSNSNGSGN NNNSSSNASD 780
781 NSGATVIYLS PEKREQRCNG GGGGGSAGNE MNRWSNPVLD RLTGGICSSN SAQPSSQQSH 840
841 QQQPHPSQQQ QQQRRLPAAP VCSSAALRNE SYYMATNDNW SPAQMHLMTA HGNTALLIED 900
901 DAEPMSPTLP ARPHDGQHCN TSSANPSQNL AVGNQGPPIN TVPGTPSTSS NFDATYSSQL 960
961 SLETNSGVEH FSRDALGRRS ISEKHHAALD ARETGTYQRN KKLREERERE RRIQLTKSAV1020
1021 YGGSIESLTA RIASANAQFS GYKHAKTASS IEQRETQQQL AAAEAEARDQ LGDLGPSLGM1080
1081 KKSSSLESLQ TMVQELQMSD EPRGHQALRA PRGRGREDSL RAAVVSEPDA SKPRKTWLLE1140
1141 DGDHEGGFAS QRNGPFQSSL NDGKHGCKSS RAKKPSILRG IGHMFRFGKN RKDGVVPVDN1200
1201 YAVNISPPTS VVSTATSPQL QQQQQQQLQQ HQQQQQIPTA ALAALERNGK PPAYQPPPPL1260
1261 PAPNGVGSNG IHQNDIFNHR YQHYANYEDL HQQHQQHQIS RRHQHYHSQR SARSQDVSMH1320
1321 STSSGSQPGS LAQPQAQSNG VRPMSSYYEY ETVQQQRVGS IKHSHSSSAT SSSSSPINVP1380
1381 HWKAAAMNGY SPASLNSSAR SRGPFVTQVT IREQSSGGIP AHLLQQHQQQ QLQQQQPTYQ1440
1441 TVQKMSGPSQ YGSAAGSQPH ASKV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
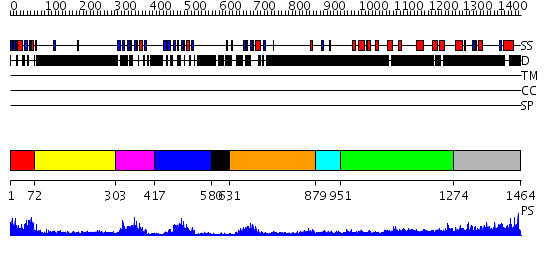
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] | 23.045757 | No description for 2ns5A was found. | |
| 2 | View Details | [72..302] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [303..416] | 18.154902 | Solution structure of the alternatively spliced PDZ2 domain (PDZ2b) of PTP-Bas (hPTP1E) | |
| 4 | View Details | [417..579] | 22.30103 | The crystal structure of the 13th PDZ domain of MPDZ | |
| 5 | View Details | [580..630] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [631..878] | 3.69 | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | |
| 7 | View Details | [879..950] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [951..1273] | 22.0 | Psd-95; Guanylate kinase-like domain of Psd-95 | |
| 9 | View Details | [1274..1464] | 2.108991 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)