













| General Information: |
|
| Name(s) found: |
CG8939-PA /
FBpp0073943
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 817 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[ISS]
|
| Biological Process: |
rRNA methylation
[IEA]
rRNA processing [ISS] |
| Molecular Function: |
nucleic acid binding
[IEA]
rRNA methyltransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKKTKVGKT RKDKFYQLAK ETGLRSRAAF KLIQLNRKFG FLQQSQVCLD LCAAPGGWMQ 60
61 VAKQNMPVSS IVIGVDLFPI RPIAGCIGLV EDITTEKCRQ SLTKELQSWK ADVVLHDGAP 120
121 NVGRNWLYDA YQQICLTLNA LKLSTQFLRN GGWFVTKVFR SKDYNALLWV LKQLFKKVHA 180
181 TKPSASRKES AEIFVVCQGY LAPDHIDPRL LDSKYVFEEL DLDAKKKSSL LHPEKQKRIK 240
241 AEGYTAQDIA LRNDLAATEF LKAENALAAL QGIGSIRIDD QRIANHKKTT PEILECCKDL 300
301 KVLGRKDIKG LVQWWKDVRE LFVEKETPLV VGNAADEKPK PLTQAEIEDM EDAELQTQIE 360
361 TIVEEEKKDL KRKRKKTLKT KAKLHEKMNL NMVIKGDDGP VEEMEHEIFD LKNIRTSAEL 420
421 DELLDVQPDF AVDESQPEET KLPKYKYFDK DEGNINDDLN YDDDNDEEQS EASDDEDQIN 480
481 DKQGLGLSDD DNDEGNAKGK NKRKKRSENP LIKSNDFRDK NTKREQRVQL WYEKDVLQNI 540
541 QSDDDEETYD LNNLAKEFKA KGVAVLGEST LTPDANEEVV QGKKAKRRAR HETATKDSSS 600
601 ESSESEDDAN EDSDGEQPAA GTNAKKVRLS ENEMALGALL TRSKKTRRDL IDAAWNRYAF 660
661 NDENVPVWFK LDEDEHMTKP TPVPKELTAE YQRKLQEVNV RPIKKVMEAK ARKHRRATKR 720
721 LAKAKKMAEK IMENSDATNH EKSKQLKKVY KKAQEKKKEV TYVVAKKQTA ARRARRPAGV 780
781 KGRYKVVDPR EKKDKRSMDA KKRREKGSKG RKGKGRK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
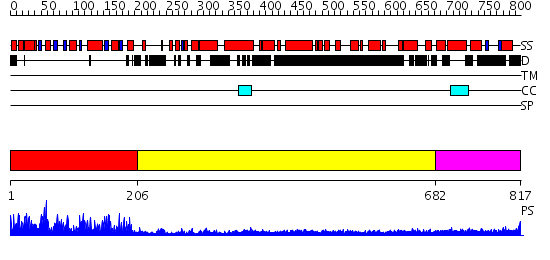
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..205] | 79.0 | RNA methyltransferase FtsJ | |
| 2 | View Details | [206..681] | 28.39794 | Heavy meromyosin subfragment | |
| 3 | View Details | [682..817] | 4.39794 | No description for 2dfsA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)