













| General Information: |
|
| Name(s) found: |
FBpp0309513
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1201 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
proteolysis
[IEA]
|
| Molecular Function: |
zinc ion binding
[IEA]
metallocarboxypeptidase activity [IEA] purine nucleotide binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEREREAMS ASLSRLLRTL SSASMSRRHC PLALLQPIPL HQSEIALGSA SSGGSAERCC 60
61 SSCSDDSNNN ERCKSAERGT AADEVGEEGR EEVELESKED AVEGEQDELE LKRDSSTVNV 120
121 NVVRANFKCN DMECSLVLGD YVNGFLGSFL SKGLKTNQLV VNTDEKTLRP VARLKEPRDL 180
181 FALPKDKDND CSQQAPRWPV ECQVIEERII HIPYVPAVPE PLNAPTGNEL KPRPVGEENG 240
241 IVVFSYSPIS AVNYEKPKAK KEDDESSDES DYSSDSRQDS TPPSRSTPMR LAGGGECAPG 300
301 KLNSVSKMIN NVDNRSTSAS LDDNDDYYDD EYDTSGGYCG GSGEAKEKAI IKDLIEAKKK 360
361 RYQEEAEVTP VGGGTARNSR AASRSEASKD ASDIDDIWKN NTSEYKPASP RYVLSQFGKN 420
421 VTKAVIDRIV DHEDLVPPSG SQKVSNQFAV GPVKLPKTNI ARLEVAFERS ACQDRSKSRL 480
481 KKKMSSSRRR RESTSDEDSS SSSLGNEEDE EVDDSDSDAE NTGSGIGLRR ILGSYSARLA 540
541 GGAEKCPCPG SGSVSDTETL VGDESRSRLA GSNGLHQQDV VCSRNRQAQS RSPLRSVPHT 600
601 HAATVAAAAA ALARGRNRDK DGCLGGKDEK NMGMGVGTTG TSSMGTRTSS PVRNNVFRLS 660
661 KDQHILLNSM TSQETTGTLI SSTSTNQTTT TTNSSQSFLC SDLPQAQFSR SAVGGARFMT 720
721 NCHPMNPEEY DGLEFESRFE SGNLAKAVQI TPTYYELYLR PDLYTSRSKQ WFYFRVRRTR 780
781 RKMLYRFSIV NLVKSDSLYN DGMQPVMYST LGAKEKSEGW RRCGDNICYY RNDDESASSS 840
841 ANEDDEDNST YTLTFTIEFE HDDDTVFFAH SYPYTYSDLQ DYLMEIQRHP VKSKFCKLRL 900
901 LCRTLAGNNV YYLTVTAPSS NEENMRRKKS IVVSARVHPS ETPASWMMKG LMDFITGDTT 960
961 VAKRLRHKFI FKLVPMLNPD GVIVGNTRNS LTGKDLNRQY RTVIRETYPS IWYTKAMIRR1020
1021 LIEECGVAMY CDMHAHSRKH NIFIYGCENK RNPEKKLTEQ VFPLMLHKNS ADRFSFESCK1080
1081 FKIQRSKEGT GRIVVWMLGI TNSYTIEASF GGSSLGSRKG THFNTQDYEH MGRAFCETLL1140
1141 DYCDENPNKV KRHAKLFKQI KKIRKREKRE QKALKLQKMA DQGCIMNDKL KVKRSVEKSV1200
1201 S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
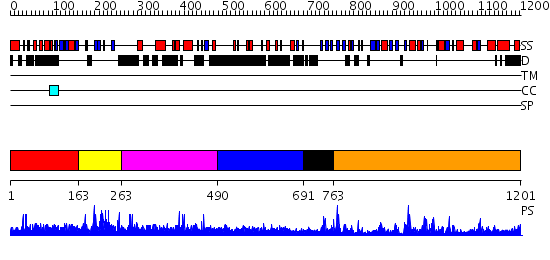
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..162] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [163..262] | 2.006965 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [263..489] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [490..690] | 6.066994 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [691..762] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [763..1201] | 70.0 | Carboxypeptidase B; Procarboxypeptidase B |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.64 |
Source: Reynolds et al. (2008)