













| General Information: |
|
| Name(s) found: |
gi|7291081
[NCBI NR]
gi|24640867 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 235 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS]
protein complex [IPI] histone acetyltransferase complex [IEA] |
| Biological Process: |
regulation of mitosis
[IGI][IMP]
hemopoiesis [TAS] smoothened signaling pathway [IMP][NAS] regulation of transcription, DNA-dependent [IEA] histone H4-K12 acetylation [IMP] histone H4-K8 acetylation [IMP] R7 cell differentiation [IMP] R3/R4 cell fate commitment [IMP] histone H3-K18 acetylation [IDA] positive regulation of transcription from RNA polymerase II promoter [IGI][IMP] regulation of transcription [IEA] histone H3-K27 acetylation [IDA] neurotransmitter secretion [IMP] synapse assembly [IMP] circadian regulation of gene expression [IMP] compound eye morphogenesis [IMP] locomotor rhythm [IMP] Wnt receptor signaling pathway [NAS] |
| Molecular Function: |
cAMP response element binding protein binding
[TAS]
histone acetyltransferase activity [IDA] zinc ion binding [IEA] protein binding [IPI] transcription coactivator activity [IDA][IGI][ISS] transcription factor binding [IPI] histone acetyltransferase activity (H3-K18 specific) [IDA] histone acetyltransferase activity (H3-K27 specific) [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHACVCCCST NNNCNNENND GSSSSSSNHN NNDSSISSSS KNKLQLKKRR AEVEEEEEVL 60
61 VKRTQDKNKH QEEQEQEEHQ AVEKLSAGQS ASGGDNDLNG AATRRRWWLR DPPGSAEAAE 120
121 GEGKGEGVEV GVGAEGSAGT GPTTWWWSAP SVDVAVTIAP ATLLLSLQRQ LAKELSKSTT 180
181 TTATRTTTEA TSLAIKSGAA ALISSLSASI ANIIDYKIPK HQYQNQIPIL ASHAF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
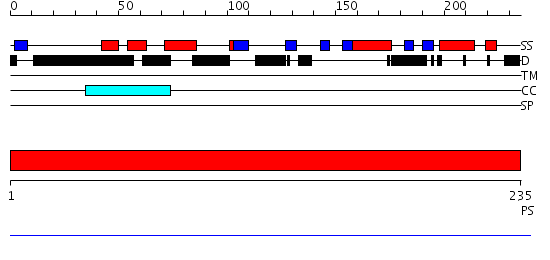
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..235] | 1.002926 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.75 |
Source: Reynolds et al. (2008)