













| General Information: |
|
| Name(s) found: |
l(1)G0148-PA /
FBpp0070911
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 700 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
autophagic cell death
[IEP]
salivary gland cell autophagic cell death [IEP] protein amino acid phosphorylation [IEA][NAS] |
| Molecular Function: |
ATP binding
[IEA]
receptor signaling protein serine/threonine kinase activity [NAS] protein serine/threonine kinase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADRVMCQEL ATPGSHYRRP KTGVPSTNVV YANGGGSWRA APRQTQRLPR QTVQQYATGT 60
61 PATGSAGRHS GRAFATAGAT EEQTVEFNGA AVMGLPATPR TTKQARNKSE EAINDLLTRI 120
121 PDIGKLFDVH SRIGNGTFST VLLGTLRRES HLPDSLRRKF AIKHHIPTSH PDRIMKELQC 180
181 MTKMGGKENV VGIHCCMRYD ASAAFVMPFM AHDRFQDFYT RMDVPEIRQY MRNLLVALRH 240
241 VHKFDVIHRD VKPSNFLYNR RRREFLLVDF GLAQHVNPPA ARSSGSAAAI AAANNKNNNN 300
301 NNNNNSKRPR ERESKGDVQQ IALDAGLGGA VKRMRLHEES NKMPLKPVND IAPSDAPEQS 360
361 VDGSNHVQPQ LVQQEQQQLQ PQQQQQQQQQ QQQSQQQQQP QQQSQQQHPQ RQPQLAQMDQ 420
421 TASTPSGSKY NTNRNVSAAA ANNAKCVCFA NPSVCLNCLM KKEVHASRAG TPGYRPPEVL 480
481 LKYPDQTTAV DVWAAGVIFL SIMSTVYPFF KAPNDFIALA EIVTIFGDQA IRKTALALDR 540
541 MITLSQRSRP LNLRKLCLRF RYRSVFSDAK LLKSYESVDG SCEVCRNCDQ YFFNCLCEDS 600
601 DYLTEPLDAY ECFPPSAYDL LDRLLEINPH KRITAEEALK HPFFTAAEEA EQTEQDQLAN 660
661 GTPRKMRRQR YQSHRTVAAS QEQVKQQVAL DLQQAAINKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
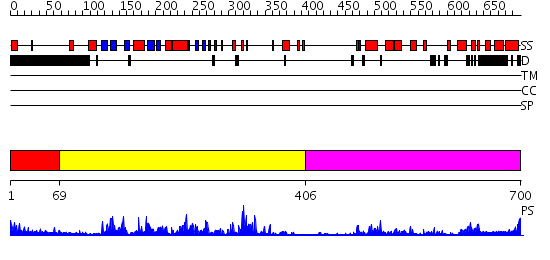
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..68] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [69..405] | 64.69897 | No description for 2gnfA was found. | |
| 3 | View Details | [406..700] | 7.25 | Crystal Structure of Human CDK7 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)