













| General Information: |
|
| Name(s) found: |
Parg-PA /
FBpp0070560
[FlyBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 768 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleoplasm
[IDA]
|
| Biological Process: |
carbohydrate metabolic process
[IMP]
response to heat [IMP] phagocytosis, engulfment [IMP] regulation of RNA splicing [IMP] regulation of histone acetylation [IMP] chromatin silencing [IMP] maintenance of protein location in nucleus [IMP] |
| Molecular Function: |
poly(ADP-ribose) glycohydrolase activity
[IMP][ISS][NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQEFRSHLIF PIFQKVYQST ANRRRASASV LTNRLGKALC LNCARMSKSP DGGISEIETE 60
61 EEPENLANSL DDSWRGVSME AIHRNRQPFE LENLPPVTAG NLHRVMYQLP IRETPPRPYK 120
121 SPGKWDSEHV RLPCAPESKY PRENPDGSTT IDFRWEMIER ALLQPIKTCE ELQAAIISYN 180
181 TTYRDQWHFR ALHQLLDEEL DESETRVFFE DLLPRIIRLA LRLPDLIQSP VPLLKHHKNA 240
241 SLSLSQQQIS CLLANAFLCT FPRRNTLKRK SEYSTFPDIN FNRLYQSTGP AVLEKLKCIM 300
301 HYFRRVCPTE RDASNVPTGV VTFVRRSGLP EHLIDWSQSA APLGDVPLHV DAEGTIEDEG 360
361 IGLLQVDFAN KYLGGGVLGH GCVQEEIRFV ICPELLVGKL FTECLRPFEA LVMLGAERYS 420
421 NYTGYAGSFE WSGNFEDSTP RDSSGRRQTA IVAIDALHFA QSHHQYREDL MERELNKAYI 480
481 GFVHWMVTPP PGVATGNWGC GAFGGDSYLK ALLQLMVCAQ LGRPLAYYTF GNVEFRDDFH 540
541 EMWLLFRNDG TTVQQLWSIL RSYSRLIKEK SSKEPRENKA SKKKLYDFIK EELKKVRDVP 600
601 GEGASAEAGS SRVAGLGEGK SETSAKSSPE LNKQPARPQI TITQQSTDLL PAQLSQDNSN 660
661 SSEDQALLML SDDEEANAMM EAASLEAKSS VEISNSSTTS KTSSTATKSM GSGGRQLSLL 720
721 EMLDTHYEKG SASKRPRKSP NCSKAEGSAK SRKEIDVTDK DEKDDIVD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
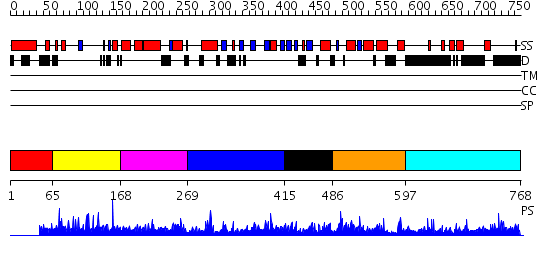
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..64] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [65..167] | 2.037979 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [168..268] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [269..414] | 1.073957 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [415..485] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [486..596] | 11.082993 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [597..768] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)